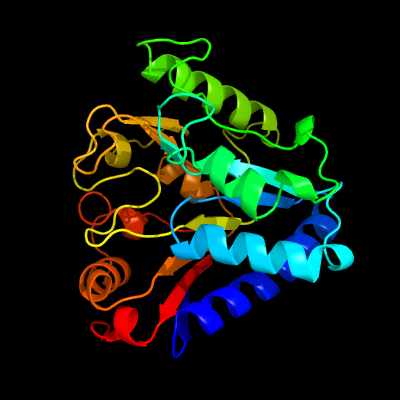
| 1 | c3icoA_
|
|
 |
100.0 |
100 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: crystal structure of 6-phosphogluconolactonase from mycobacterium2 tuberculosis
|
|
|
|
| 2 | c3oc6A_
|
|
 |
100.0 |
66 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: crystal structure of 6-phosphogluconolactonase from mycobacterium2 smegmatis, apo form
|
|
|
|
| 3 | c2j0eA_
|
|
 |
100.0 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: three dimensional structure and catalytic mechanism of 6-2 phosphogluconolactonase from trypanosoma brucei
|
|
|
|
| 4 | c3hn6D_
|
|
 |
100.0 |
22 |
PDB header:isomerase
Chain: D: PDB Molecule:glucosamine-6-phosphate deaminase;
PDBTitle: crystal structure of glucosamine-6-phosphate deaminase from borrelia2 burgdorferi
|
|
|
|
| 5 | d1ne7a_
|
|
 |
100.0 |
24 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:NagB-like |
|
|
|
| 6 | c1y89B_
|
|
 |
100.0 |
31 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:devb protein;
PDBTitle: crystal structure of devb protein
|
|
|
|
| 7 | c3cssA_
|
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: crystal structure of 6-phosphogluconolactonase from leishmania2 guyanensis
|
|
|
|
| 8 | d1fsfa_
|
|
 |
100.0 |
22 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:NagB-like |
|
|
|
| 9 | c6nbgD_
|
|
 |
100.0 |
22 |
PDB header:unknown function
Chain: D: PDB Molecule:glucosamine-6-phosphate deaminase;
PDBTitle: 2.05 angstrom resolution crystal structure of hypothetical protein2 kp1_5497 from klebsiella pneumoniae.
|
|
|
|
| 10 | c1pbtA_
|
|
 |
100.0 |
29 |
PDB header:hydrolase, oxidoreductase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: the crystal structure of tm1154, oxidoreductase, sol/devb2 family from thermotoga maritima
|
|
|
|
| 11 | d1vl1a_
|
|
 |
100.0 |
29 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:NagB-like |
|
|
|
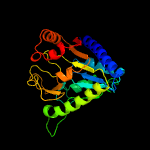
| 12 | c3lwdA_
|
|
 |
100.0 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: crystal structure of putative 6-phosphogluconolactonase (yp_574786.1)2 from chromohalobacter salexigens dsm 3043 at 1.88 a resolution
|
|
|
|
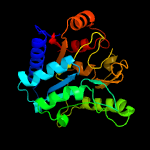
| 13 | c3e15D_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: D: PDB Molecule:glucose-6-phosphate 1-dehydrogenase;
PDBTitle: 6-phosphogluconolactonase from plasmodium vivax
|
|
|
|
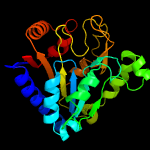
| 14 | c3nwpA_
|
|
 |
100.0 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phosphogluconolactonase;
PDBTitle: crystal structure of a 6-phosphogluconolactonase (sbal_2240) from2 shewanella baltica os155 at 1.40 a resolution
|
|
|
|
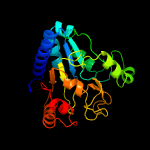
| 15 | c3lhiA_
|
|
 |
100.0 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative 6-phosphogluconolactonase;
PDBTitle: crystal structure of putative 6-phosphogluconolactonase(yp_207848.1)2 from neisseria gonorrhoeae fa 1090 at 1.33 a resolution
|
|
|
|
| 16 | c2bkxB_
|
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:glucosamine-6-phosphate deaminase;
PDBTitle: structure and kinetics of a monomeric glucosamine-6-2 phosphate deaminase: missing link of the nagb superfamily
|
|
|
|
| 17 | c2ri0B_
|
|
 |
100.0 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:glucosamine-6-phosphate deaminase;
PDBTitle: crystal structure of glucosamine 6-phosphate deaminase (nagb) from s.2 mutans
|
|
|
|
| 18 | c4oqqA_
|
|
 |
99.8 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:deoxyribonucleoside regulator;
PDBTitle: structure of the effector-binding domain of deoxyribonucleoside2 regulator deor from bacillus subtilis
|
|
|
|
| 19 | d2gnpa1
|
|
 |
99.7 |
11 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
|
|
|
| 20 | c2w48D_
|
|
 |
99.6 |
14 |
PDB header:transcription
Chain: D: PDB Molecule:sorbitol operon regulator;
PDBTitle: crystal structure of the full-length sorbitol operon2 regulator sorc from klebsiella pneumoniae
|
|
|
|
| 21 | d2o0ma1 |
|
not modelled |
99.5 |
15 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
|
|
| 22 | c2o0mA_ |
|
not modelled |
99.5 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, sorc family;
PDBTitle: the crystal structure of the putative sorc family transcriptional2 regulator from enterococcus faecalis
|
|
|
| 23 | d3efba1 |
|
not modelled |
99.5 |
14 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
|
|
| 24 | c3nzeB_ |
|
not modelled |
99.4 |
17 |
PDB header:transcription regulator
Chain: B: PDB Molecule:putative transcriptional regulator, sugar-binding family;
PDBTitle: the crystal structure of a domain of a possible sugar-binding2 transcriptional regulator from arthrobacter aurescens tc1.
|
|
|
| 25 | c4r9nA_ |
|
not modelled |
99.4 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:lmo0547 protein;
PDBTitle: deor family transcriptional regulator from listeria monocytogenes.
|
|
|
| 26 | c4go1A_ |
|
not modelled |
98.7 |
22 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator lsrr;
PDBTitle: crystal structure of full length transcription repressor lsrr from e.2 coli.
|
|
|
| 27 | d2okga1 |
|
not modelled |
98.5 |
14 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
|
|
| 28 | c3kv1A_ |
|
not modelled |
97.5 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional repressor;
PDBTitle: crystal structure of putative sugar-binding domain of transcriptional2 repressor from vibrio fischeri
|
|
|
| 29 | d2r5fa1 |
|
not modelled |
97.4 |
16 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
|
|
| 30 | c2e21A_ |
|
not modelled |
91.9 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:trna(ile)-lysidine synthase;
PDBTitle: crystal structure of tils in a complex with amppnp from aquifex2 aeolicus.
|
|
|
| 31 | d1zuna1 |
|
not modelled |
86.9 |
8 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:PAPS reductase-like |
|
|
| 32 | c1ni5A_ |
|
not modelled |
81.5 |
14 |
PDB header:cell cycle
Chain: A: PDB Molecule:putative cell cycle protein mesj;
PDBTitle: structure of the mesj pp-atpase from escherichia coli
|
|
|
| 33 | d1wy5a1 |
|
not modelled |
77.0 |
14 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:PP-loop ATPase |
|
|
| 34 | c3vrhA_ |
|
not modelled |
76.2 |
9 |
PDB header:rna binding protein
Chain: A: PDB Molecule:putative uncharacterized protein ph0300;
PDBTitle: crystal structure of ph0300
|
|
|
| 35 | c5udwB_ |
|
not modelled |
73.4 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:lactate racemization operon protein lare;
PDBTitle: lare, a sulfur transferase involved in synthesis of the cofactor for2 lactate racemase, in complex with nickel
|
|
|
| 36 | c3a2kB_ |
|
not modelled |
72.8 |
15 |
PDB header:ligase/rna
Chain: B: PDB Molecule:trna(ile)-lysidine synthase;
PDBTitle: crystal structure of tils complexed with trna
|
|
|
| 37 | c2oq2B_ |
|
not modelled |
66.6 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:phosphoadenosine phosphosulfate reductase;
PDBTitle: crystal structure of yeast paps reductase with pap, a product complex
|
|
|
| 38 | d1ni5a1 |
|
not modelled |
56.5 |
14 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:PP-loop ATPase |
|
|
| 39 | c2goyC_ |
|
not modelled |
53.5 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:adenosine phosphosulfate reductase;
PDBTitle: crystal structure of assimilatory adenosine 5'-2 phosphosulfate reductase with bound aps
|
|
|
| 40 | c1zunA_ |
|
not modelled |
52.1 |
8 |
PDB header:transferase
Chain: A: PDB Molecule:sulfate adenylyltransferase subunit 2;
PDBTitle: crystal structure of a gtp-regulated atp sulfurylase2 heterodimer from pseudomonas syringae
|
|
|
| 41 | c2q4dB_ |
|
not modelled |
48.6 |
22 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:lysine decarboxylase-like protein at5g11950;
PDBTitle: ensemble refinement of the crystal structure of a lysine2 decarboxylase-like protein from arabidopsis thaliana gene at5g11950
|
|
|
| 42 | c3uowB_ |
|
not modelled |
48.2 |
8 |
PDB header:ligase
Chain: B: PDB Molecule:gmp synthetase;
PDBTitle: crystal structure of pf10_0123, a gmp synthetase from plasmodium2 falciparum
|
|
|
| 43 | d1wxia1 |
|
not modelled |
46.5 |
9 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
|
|
| 44 | d1pjqa3 |
|
not modelled |
45.9 |
19 |
Fold:Siroheme synthase middle domains-like
Superfamily:Siroheme synthase middle domains-like
Family:Siroheme synthase middle domains-like |
|
|
| 45 | c5tw7E_ |
|
not modelled |
45.5 |
15 |
PDB header:ligase
Chain: E: PDB Molecule:gmp synthase [glutamine-hydrolyzing];
PDBTitle: crystal structure of a gmp synthase (glutamine-hydrolyzing) from2 neisseria gonorrhoeae
|
|
|
| 46 | c4xgjA_ |
|
not modelled |
43.4 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a domain of unknown function (duf1537) from2 pectobacterium atrosepticum (eca3761), target efi-511609, apo3 structure, domain swapped dimer
|
|
|
| 47 | d1gpma1 |
|
not modelled |
43.3 |
15 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
|
|
| 48 | d1ydha_ |
|
not modelled |
41.7 |
22 |
Fold:MCP/YpsA-like
Superfamily:MCP/YpsA-like
Family:MoCo carrier protein-like |
|
|
| 49 | c1gpmD_ |
|
not modelled |
39.3 |
18 |
PDB header:transferase (glutamine amidotransferase)
Chain: D: PDB Molecule:gmp synthetase;
PDBTitle: escherichia coli gmp synthetase complexed with amp and pyrophosphate
|
|
|
| 50 | c2dplA_ |
|
not modelled |
38.3 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:gmp synthase [glutamine-hydrolyzing] subunit b;
PDBTitle: crystal structure of the gmp synthase from pyrococcus horikoshii ot3
|
|
|
| 51 | c3dpiA_ |
|
not modelled |
38.0 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:nad+ synthetase;
PDBTitle: crystal structure of nad+ synthetase from burkholderia pseudomallei
|
|
|
| 52 | d3bzka5 |
|
not modelled |
37.1 |
13 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Tex RuvX-like domain-like |
|
|
| 53 | c3u7jA_ |
|
not modelled |
36.8 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from burkholderia2 thailandensis
|
|
|
| 54 | c3tqiB_ |
|
not modelled |
36.7 |
13 |
PDB header:ligase
Chain: B: PDB Molecule:gmp synthase [glutamine-hydrolyzing];
PDBTitle: structure of the gmp synthase (guaa) from coxiella burnetii
|
|
|
| 55 | d1xnga1 |
|
not modelled |
36.7 |
12 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
|
|
| 56 | c5hujB_ |
|
not modelled |
36.4 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:nh(3)-dependent nad(+) synthetase;
PDBTitle: crystal structure of nade from streptococcus pyogenes
|
|
|
| 57 | d1uj4a1 |
|
not modelled |
35.9 |
19 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:D-ribose-5-phosphate isomerase (RpiA), catalytic domain |
|
|
| 58 | c4bwvB_ |
|
not modelled |
35.2 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:phosphoadenosine-phosphosulphate reductase;
PDBTitle: structure of adenosine 5-prime-phosphosulfate reductase apr-b from2 physcomitrella patens
|
|
|
| 59 | c4xfdA_ |
|
not modelled |
33.0 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:nh(3)-dependent nad(+) synthetase;
PDBTitle: crystal structure of a nh(3)-dependent nad(+) synthetase from2 pseudomonas aeruginosa
|
|
|
| 60 | c4q16C_ |
|
not modelled |
32.5 |
18 |
PDB header:ligase
Chain: C: PDB Molecule:nh(3)-dependent nad(+) synthetase;
PDBTitle: structure of nad+ synthetase from deinococcus radiodurans
|
|
|
| 61 | c5itsD_ |
|
not modelled |
32.1 |
25 |
PDB header:hydrolase
Chain: D: PDB Molecule:cytokinin riboside 5'-monophosphate phosphoribohydrolase;
PDBTitle: crystal strcuture of log from corynebacterium glutamicum
|
|
|
| 62 | c3sftA_ |
|
not modelled |
31.9 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:chemotaxis response regulator protein-glutamate
PDBTitle: crystal structure of thermotoga maritima cheb methylesterase catalytic2 domain
|
|
|
| 63 | d1q15a1 |
|
not modelled |
31.7 |
19 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
|
|
| 64 | d1t35a_ |
|
not modelled |
31.3 |
19 |
Fold:MCP/YpsA-like
Superfamily:MCP/YpsA-like
Family:MoCo carrier protein-like |
|
|
| 65 | c5zbjA_ |
|
not modelled |
29.6 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative cytokinin riboside 5'-monophosphate
PDBTitle: crystal strcuture of type-i log from pseudomonas aeruginosa pao1
|
|
|
| 66 | c2o8vA_ |
|
not modelled |
29.3 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phosphoadenosine phosphosulfate reductase;
PDBTitle: paps reductase in a covalent complex with thioredoxin c35a
|
|
|
| 67 | d1xhoa_ |
|
not modelled |
26.0 |
21 |
Fold:Bacillus chorismate mutase-like
Superfamily:YjgF-like
Family:Chorismate mutase |
|
|
| 68 | c2q4oA_ |
|
not modelled |
25.3 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein at2g37210/t2n18.3;
PDBTitle: ensemble refinement of the crystal structure of a lysine2 decarboxylase-like protein from arabidopsis thaliana gene at2g37210
|
|
|
| 69 | d2q4oa1 |
|
not modelled |
25.3 |
21 |
Fold:MCP/YpsA-like
Superfamily:MCP/YpsA-like
Family:MoCo carrier protein-like |
|
|
| 70 | c1xhoB_ |
|
not modelled |
24.2 |
21 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:chorismate mutase;
PDBTitle: chorismate mutase from clostridium thermocellum cth-682
|
|
|
| 71 | d1kqpa_ |
|
not modelled |
24.2 |
13 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
|
|
| 72 | c6c6kD_ |
|
not modelled |
23.9 |
43 |
PDB header:rna binding protein/rna
Chain: D: PDB Molecule:interferon-induced protein with tetratricopeptide repeats
PDBTitle: structural basis for preferential recognition of cap 0 rna by a human2 ifit1-ifit3 protein complex
|
|
|
| 73 | c1uj6A_ |
|
not modelled |
23.3 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: crystal structure of thermus thermophilus ribose-5-phosphate isomerase2 complexed with arabinose-5-phosphate
|
|
|
| 74 | c2vzaD_ |
|
not modelled |
23.1 |
14 |
PDB header:cell adhesion
Chain: D: PDB Molecule:cell filamentation protein;
PDBTitle: type iv secretion system effector protein bepa
|
|
|
| 75 | c4gmkB_ |
|
not modelled |
22.1 |
11 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose 5-phosphate isomerase from the probiotic2 bacterium lactobacillus salivarius ucc118
|
|
|
| 76 | d1ct9a1 |
|
not modelled |
21.4 |
14 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
|
|
| 77 | c1x3lA_ |
|
not modelled |
21.3 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:hypothetical protein ph0495;
PDBTitle: crystal structure of the ph0495 protein from pyrococccus horikoshii2 ot3
|
|
|
| 78 | c4x84C_ |
|
not modelled |
21.1 |
16 |
PDB header:isomerase
Chain: C: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from pseudomonas2 aeruginosa
|
|
|
| 79 | d1jgta1 |
|
not modelled |
19.9 |
20 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
|
|
| 80 | d1yzya1 |
|
not modelled |
19.7 |
14 |
Fold:YgbK-like
Superfamily:YgbK-like
Family:YgbK-like |
|
|
| 81 | c3q4gA_ |
|
not modelled |
19.6 |
13 |
PDB header:ligase
Chain: A: PDB Molecule:nh(3)-dependent nad(+) synthetase;
PDBTitle: structure of nad synthetase from vibrio cholerae
|
|
|
| 82 | d2jm6b1 |
|
not modelled |
19.6 |
8 |
Fold:Toxins' membrane translocation domains
Superfamily:Bcl-2 inhibitors of programmed cell death
Family:Bcl-2 inhibitors of programmed cell death |
|
|
| 83 | d2fvta1 |
|
not modelled |
19.5 |
25 |
Fold:MTH938-like
Superfamily:MTH938-like
Family:MTH938-like |
|
|
| 84 | c3e59A_ |
|
not modelled |
19.5 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:pyoverdine biosynthesis protein pvca;
PDBTitle: crystal structure of the pvca (pa2254) protein from pseudomonas2 aeruginosa
|
|
|
| 85 | c5zi9B_ |
|
not modelled |
19.3 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:cytokinin riboside 5'-monophosphate phosphoribohydrolase;
PDBTitle: crystal strcuture of type-ii log from streptomyces coelicolor a3
|
|
|
| 86 | d1dbfa_ |
|
not modelled |
18.6 |
12 |
Fold:Bacillus chorismate mutase-like
Superfamily:YjgF-like
Family:Chorismate mutase |
|
|
| 87 | c2f8mB_ |
|
not modelled |
18.6 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: ribose 5-phosphate isomerase from plasmodium falciparum
|
|
|
| 88 | c2gaaA_ |
|
not modelled |
18.2 |
9 |
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical 39.9 kda protein;
PDBTitle: crystal structure of yfh7 from saccharomyces cerevisiae: a2 putative p-loop containing kinase with a circular3 permutation.
|
|
|
| 89 | c3l7oB_ |
|
not modelled |
17.8 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from streptococcus2 mutans ua159
|
|
|
| 90 | c3fiuD_ |
|
not modelled |
17.5 |
9 |
PDB header:ligase
Chain: D: PDB Molecule:nh(3)-dependent nad(+) synthetase;
PDBTitle: structure of nmn synthetase from francisella tularensis
|
|
|
| 91 | c6gwjK_ |
|
not modelled |
16.6 |
21 |
PDB header:rna binding protein
Chain: K: PDB Molecule:probable trna n6-adenosine threonylcarbamoyltransferase;
PDBTitle: protein complex
|
|
|
| 92 | c1y8qA_ |
|
not modelled |
16.5 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:ubiquitin-like 1 activating enzyme e1a;
PDBTitle: sumo e1 activating enzyme sae1-sae2-mg-atp complex
|
|
|
| 93 | d2a7ya1 |
|
not modelled |
16.4 |
32 |
Fold:SH3-like barrel
Superfamily:Cell growth inhibitor/plasmid maintenance toxic component
Family:Rv2302-like |
|
|
| 94 | c2a7yA_ |
|
not modelled |
16.4 |
32 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein rv2302/mt2359;
PDBTitle: solution structure of the conserved hypothetical protein2 rv2302 from the bacterium mycobacterium tuberculosis
|
|
|
| 95 | c1lk5C_ |
|
not modelled |
16.3 |
17 |
PDB header:isomerase
Chain: C: PDB Molecule:d-ribose-5-phosphate isomerase;
PDBTitle: structure of the d-ribose-5-phosphate isomerase from2 pyrococcus horikoshii
|
|
|
| 96 | c4xfrB_ |
|
not modelled |
15.5 |
33 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a domain of unknown function (duf1537) from2 bordetella bronchiseptica (bb3215), target efi-511620, with bound3 citrate, domain swapped dimer, space group p6522
|
|
|
| 97 | c5ghaC_ |
|
not modelled |
15.3 |
18 |
PDB header:transferase/transport protein
Chain: C: PDB Molecule:sulfur transferase ttua;
PDBTitle: sulfur transferase ttua in complex with sulfur carrier ttub
|
|
|
| 98 | c5wq3A_ |
|
not modelled |
15.2 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:cytokinin riboside 5'-monophosphate phosphoribohydrolase;
PDBTitle: crystal strcuture of type-ii log from corynebacterium glutamicum
|
|
|
| 99 | d2b8na1 |
|
not modelled |
15.1 |
22 |
Fold:GckA/TtuD-like
Superfamily:GckA/TtuD-like
Family:GckA/TtuD-like |
|
|