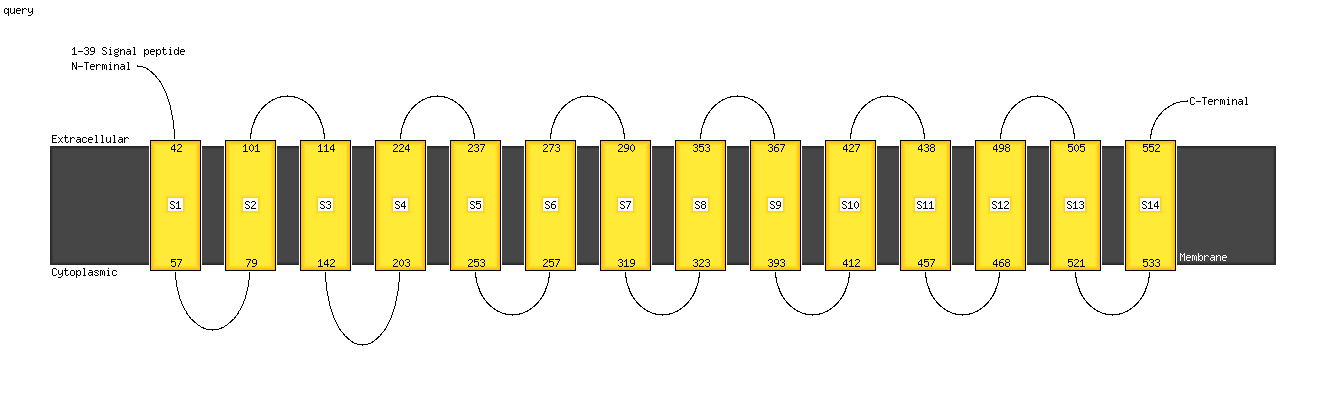
1 c5f15A_
99.0
15
PDB header: transferaseChain: A: PDB Molecule: 4-amino-4-deoxy-l-arabinose (l-ara4n) transferase;PDBTitle: crystal structure of arnt from cupriavidus metallidurans bound to2 undecaprenyl phosphate
2 c6p25A_
98.4
17
PDB header: transferaseChain: A: PDB Molecule: dolichyl-phosphate-mannose--protein mannosyltransferase 1;PDBTitle: structure of s. cerevisiae protein o-mannosyltransferase pmt1-pmt22 complex bound to the sugar donor and a peptide acceptor
3 c6p2rB_
98.1
16
PDB header: transferaseChain: B: PDB Molecule: dolichyl-phosphate-mannose--protein mannosyltransferase 2;PDBTitle: structure of s. cerevisiae protein o-mannosyltransferase pmt1-pmt22 complex bound to the sugar donor
4 c3wajA_
97.8
14
PDB header: transferaseChain: A: PDB Molecule: transmembrane oligosaccharyl transferase;PDBTitle: crystal structure of the archaeoglobus fulgidus2 oligosaccharyltransferase (o29867_arcfu) complex with zn and sulfate
5 c3rceA_
96.9
11
PDB header: transferase/peptideChain: A: PDB Molecule: oligosaccharide transferase to n-glycosylate proteins;PDBTitle: bacterial oligosaccharyltransferase pglb
6 c2kfvA_
36.0
26
PDB header: isomeraseChain: A: PDB Molecule: fk506-binding protein 3;PDBTitle: structure of the amino-terminal domain of human fk506-2 binding protein 3 / northeast structural genomics3 consortium target ht99a
7 c6adqP_
30.6
14
PDB header: electron transportChain: P: PDB Molecule: prokaryotic respiratory supercomplex associate factor 1PDBTitle: respiratory complex ciii2civ2sod2 from mycobacterium smegmatis
8 d1ynjd1
30.2
22
Fold: beta and beta-prime subunits of DNA dependent RNA-polymeraseSuperfamily: beta and beta-prime subunits of DNA dependent RNA-polymeraseFamily: RNA-polymerase beta-prime
9 c5ir6A_
26.7
23
PDB header: oxidoreductaseChain: A: PDB Molecule: bd-type quinol oxidase subunit i;PDBTitle: the structure of bd oxidase from geobacillus thermodenitrificans
10 d2r6gf1
25.9
7
Fold: MalF N-terminal region-likeSuperfamily: MalF N-terminal region-likeFamily: MalF N-terminal region-like
11 c4g7oN_
25.5
25
PDB header: transcription, transferase/dnaChain: N: PDB Molecule: dna-directed rna polymerase subunit beta';PDBTitle: crystal structure of thermus thermophilus transcription initiation2 complex containing 2 nt of rna
12 c5xj0D_
24.8
25
PDB header: transferase/transcriptionChain: D: PDB Molecule: dna-directed rna polymerase subunit beta';PDBTitle: t. thermophilus rna polymerase holoenzyme bound with gp39 and gp76
13 c5x22D_
18.9
25
PDB header: transferase/dnaChain: D: PDB Molecule: dna-directed rna polymerase subunit beta';PDBTitle: crystal structure of thermus thermophilus transcription initiation2 complex with gpa and cmpcpp
14 d1zxoa1
18.5
15
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: BadF/BadG/BcrA/BcrD-like
15 d2e74b1
16.7
16
Fold: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Superfamily: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Family: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
16 d1zbsa2
15.8
15
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: BadF/BadG/BcrA/BcrD-like
17 d1vf5b_
15.2
16
Fold: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Superfamily: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Family: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
18 d1q90d_
12.8
10
Fold: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Superfamily: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Family: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
19 d1r9fa_
11.5
25
Fold: Tombusvirus P19 core protein, VP19Superfamily: Tombusvirus P19 core protein, VP19Family: Tombusvirus P19 core protein, VP19
20 c3e1kJ_
11.5
36
PDB header: transcriptionChain: J: PDB Molecule: lactose regulatory protein lac9;PDBTitle: crystal structure of kluyveromyces lactis gal80p in complex with the2 acidic activation domain of gal4p
21 c3e1kL_
not modelled
11.5
36
PDB header: transcriptionChain: L: PDB Molecule: lactose regulatory protein lac9;PDBTitle: crystal structure of kluyveromyces lactis gal80p in complex with the2 acidic activation domain of gal4p
22 c3e1kB_
not modelled
11.5
36
PDB header: transcriptionChain: B: PDB Molecule: lactose regulatory protein lac9;PDBTitle: crystal structure of kluyveromyces lactis gal80p in complex with the2 acidic activation domain of gal4p
23 c3e1kD_
not modelled
11.5
36
PDB header: transcriptionChain: D: PDB Molecule: lactose regulatory protein lac9;PDBTitle: crystal structure of kluyveromyces lactis gal80p in complex with the2 acidic activation domain of gal4p
24 c3e1kH_
not modelled
11.5
36
PDB header: transcriptionChain: H: PDB Molecule: lactose regulatory protein lac9;PDBTitle: crystal structure of kluyveromyces lactis gal80p in complex with the2 acidic activation domain of gal4p
25 c3e1kP_
not modelled
11.5
36
PDB header: transcriptionChain: P: PDB Molecule: lactose regulatory protein lac9;PDBTitle: crystal structure of kluyveromyces lactis gal80p in complex with the2 acidic activation domain of gal4p
26 c3e1kN_
not modelled
11.5
36
PDB header: transcriptionChain: N: PDB Molecule: lactose regulatory protein lac9;PDBTitle: crystal structure of kluyveromyces lactis gal80p in complex with the2 acidic activation domain of gal4p
27 c3e1kF_
not modelled
11.5
36
PDB header: transcriptionChain: F: PDB Molecule: lactose regulatory protein lac9;PDBTitle: crystal structure of kluyveromyces lactis gal80p in complex with the2 acidic activation domain of gal4p
28 c3qqcA_
not modelled
11.1
25
PDB header: transcriptionChain: A: PDB Molecule: dna-directed rna polymerase subunit b, dna-directed rnaPDBTitle: crystal structure of archaeal spt4/5 bound to the rnap clamp domain
29 c3aoiN_
not modelled
9.4
25
PDB header: transcription, transferase/dna/rnaChain: N: PDB Molecule: dna-directed rna polymerase subunit beta';PDBTitle: rna polymerase-gfh1 complex (crystal type 2)
30 c4iuwA_
not modelled
8.8
17
PDB header: hydrolaseChain: A: PDB Molecule: neutral endopeptidase;PDBTitle: crystal structure of pepo from lactobacillus rhamnosis hn001 (dr20)
31 c3aoiI_
not modelled
8.7
25
PDB header: transcription, transferase/dna/rnaChain: I: PDB Molecule: dna-directed rna polymerase subunit beta';PDBTitle: rna polymerase-gfh1 complex (crystal type 2)
32 c2o5jN_
not modelled
8.1
25
PDB header: transferase/dna-rna hybridChain: N: PDB Molecule: dna-directed rna polymerase beta' chain;PDBTitle: crystal structure of the t. thermophilus rnap polymerase elongation2 complex with the ntp substrate analog
33 c2owrD_
not modelled
8.0
54
PDB header: hydrolaseChain: D: PDB Molecule: uracil-dna glycosylase;PDBTitle: crystal structure of vaccinia virus uracil-dna glycosylase
34 d1smyd_
not modelled
7.9
25
Fold: beta and beta-prime subunits of DNA dependent RNA-polymeraseSuperfamily: beta and beta-prime subunits of DNA dependent RNA-polymeraseFamily: RNA-polymerase beta-prime
35 c3fmtF_
not modelled
7.8
40
PDB header: replication inhibitor/dnaChain: F: PDB Molecule: protein seqa;PDBTitle: crystal structure of seqa bound to dna
36 d1xrxa1
not modelled
7.3
50
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: SeqA N-terminal domain-like
37 c1xrxD_
not modelled
7.3
50
PDB header: replication inhibitorChain: D: PDB Molecule: seqa protein;PDBTitle: crystal structure of a dna-binding protein
38 c3dwbA_
not modelled
7.0
20
PDB header: hydrolaseChain: A: PDB Molecule: endothelin-converting enzyme 1;PDBTitle: structure of human ece-1 complexed with phosphoramidon
39 c5nn7A_
not modelled
6.9
43
PDB header: hydrolaseChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: kshv uracil-dna glycosylase, apo form
40 c6eznF_
not modelled
6.7
10
PDB header: membrane proteinChain: F: PDB Molecule: dolichyl-diphosphooligosaccharide--proteinPDBTitle: cryo-em structure of the yeast oligosaccharyltransferase (ost) complex
41 c4y0lA_
not modelled
6.4
44
PDB header: membrane proteinChain: A: PDB Molecule: putative membrane protein mmpl11;PDBTitle: mycobacterial membrane protein mmpl11d2
42 c3w52A_
not modelled
6.4
30
PDB header: hydrolaseChain: A: PDB Molecule: endonuclease 2;PDBTitle: zinc-dependent bifunctional nuclease
43 c3sngA_
not modelled
6.4
30
PDB header: hydrolaseChain: A: PDB Molecule: nuclease;PDBTitle: x-ray structure of fully glycosylated bifunctional nuclease tbn1 from2 solanum lycopersicum (tomato)
44 d2hu7a1
not modelled
6.3
56
Fold: 7-bladed beta-propellerSuperfamily: Peptidase/esterase 'gauge' domainFamily: Acylamino-acid-releasing enzyme, N-terminal donain
45 c3gwqB_
not modelled
6.2
25
PDB header: lyaseChain: B: PDB Molecule: d-serine deaminase;PDBTitle: crystal structure of a putative d-serine deaminase (bxe_a4060) from2 burkholderia xenovorans lb400 at 2.00 a resolution
46 c5tw1D_
not modelled
6.2
50
PDB header: transcription activator/transferase/dnaChain: D: PDB Molecule: dna-directed rna polymerase subunit beta';PDBTitle: crystal structure of a mycobacterium smegmatis transcription2 initiation complex with rbpa
47 c2o5iD_
not modelled
6.1
25
PDB header: transferase/dna-rna hybridChain: D: PDB Molecule: dna-directed rna polymerase beta' chain;PDBTitle: crystal structure of the t. thermophilus rna polymerase elongation2 complex
48 c5z3dA_
not modelled
6.1
20
PDB header: hydrolaseChain: A: PDB Molecule: glycoside hydrolase 15-related protein;PDBTitle: glycosidase f290y
49 c1qfqB_
not modelled
6.0
55
PDB header: transcription/rnaChain: B: PDB Molecule: 36-mer n-terminal peptide of the n protein;PDBTitle: bacteriophage lambda n-protein-nutboxb-rna complex
50 d1ak0a_
not modelled
5.8
10
Fold: Phospholipase C/P1 nucleaseSuperfamily: Phospholipase C/P1 nucleaseFamily: P1 nuclease
51 c4qiwA_
not modelled
5.7
25
PDB header: transcriptionChain: A: PDB Molecule: dna-directed rna polymerase;PDBTitle: crystal structure of euryarchaeal rna polymerase from thermococcus2 kodakarensis
52 c6iu3A_
not modelled
5.7
9
PDB header: metal transportChain: A: PDB Molecule: vit1;PDBTitle: crystal structure of iron transporter vit1 with zinc ions
53 d1dmta_
not modelled
5.7
14
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Neutral endopeptidase (neprilysin)
54 c2mckA_
not modelled
5.6
37
PDB header: hydrolaseChain: A: PDB Molecule: polyprotein;PDBTitle: backbone 1h, 13c, and 15n chemical shift assignments for murine2 norovirus cr6 ns1/2 protein
55 c2mmuA_
not modelled
5.6
40
PDB header: cell cycleChain: A: PDB Molecule: cell division protein crga;PDBTitle: structure of crga, a cell division structural and regulatory protein2 from mycobacterium tuberculosis, in lipid bilayers
56 c5lmxA_
not modelled
5.4
25
PDB header: transcriptionChain: A: PDB Molecule: dna-directed rna polymerase i subunit rpa190;PDBTitle: monomeric rna polymerase i at 4.9 a resolution
57 c1zzaA_
not modelled
5.3
21
PDB header: membrane proteinChain: A: PDB Molecule: stannin;PDBTitle: solution nmr structure of the membrane protein stannin
58 c5fbfA_
not modelled
5.2
30
PDB header: hydrolaseChain: A: PDB Molecule: nuclease s1;PDBTitle: s1 nuclease from aspergillus oryzae in complex with two molecules of2 2'-deoxycytidine-5'-monophosphate
59 c1i6hA_
not modelled
5.2
25
PDB header: transcription/dna-rna hybridChain: A: PDB Molecule: dna-directed rna polymerase ii largest subunit;PDBTitle: rna polymerase ii elongation complex
60 c5fjaA_
not modelled
5.1
38
PDB header: transcriptionChain: A: PDB Molecule: dna-directed rna polymerase iii subunit rpc1;PDBTitle: cryo-em structure of yeast rna polymerase iii at 4.7 a
61 d1alna2
not modelled
5.1
20
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase