1 c2kptA_
82.2
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative secreted protein;PDBTitle: solution nmr structure of the n-terminal domain of cg24962 protein from corynebacterium glutamicum. northeast3 structural genomics consortium target cgr26a
2 c5dboA_
47.7
16
PDB header: translationChain: A: PDB Molecule: translation initiation factor eif-2b-like protein;PDBTitle: crystal structure of the tetrameric eif2b-beta2-delta2 complex from c.2 thermophilum
3 c6hwhT_
39.9
14
PDB header: electron transportChain: T: PDB Molecule: uncharacterized protein msmeg_4692/msmei_4575;PDBTitle: structure of a functional obligate respiratory supercomplex from2 mycobacterium smegmatis
4 c2kw7A_
38.7
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved domain protein;PDBTitle: solution nmr structure of the n-terminal domain of protein pg_03612 from p.gingivalis, northeast structural genomics consortium target3 pgr37a
5 c4zquA_
37.9
8
PDB header: toxinChain: A: PDB Molecule: cdia-ct toxin, conserved domain protein;PDBTitle: cdia-ct/cdii toxin and immunity complex from yersinia2 pseudotuberculosis
6 c5anpB_
35.6
9
PDB header: unknown functionChain: B: PDB Molecule: ba41;PDBTitle: crystal structure of the ba41 protein from bizionia argentinensis
7 c3wrbB_
33.5
15
PDB header: oxidoreductaseChain: B: PDB Molecule: gallate dioxygenase;PDBTitle: crystal structure of the anaerobic h124f desb-gallate complex
8 c2jx5A_
25.2
21
PDB header: ribosomal proteinChain: A: PDB Molecule: glub(s27a);PDBTitle: solution structure of the ubiquitin domain n-terminal to2 the s27a ribosomal subunit of giardia lamblia
9 c2rmzA_
23.9
26
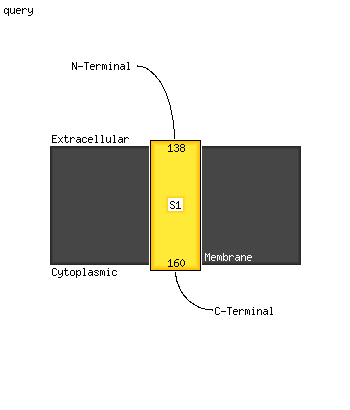
PDB header: cell adhesionChain: A: PDB Molecule: integrin beta-3;PDBTitle: bicelle-embedded integrin beta3 transmembrane segment
10 c4wpgA_
23.2
15
PDB header: oxidoreductaseChain: A: PDB Molecule: dtdp-4-dehydrorhamnose reductase;PDBTitle: group a streptococcus gaca is an essential dtdp-4-dehydrorhamnose2 reductase (rmld)
11 c2x3dC_
23.1
21
PDB header: unknown functionChain: C: PDB Molecule: sso6206;PDBTitle: crystal structure of sso6206 from sulfolobus solfataricus p2
12 c5vxsF_
22.0
13
PDB header: lyaseChain: F: PDB Molecule: citrate lyase subunit beta-like protein, mitochondrial;PDBTitle: crystal structure analysis of human clybl in apo form
13 d1gjja1
21.8
25
Fold: LEM/SAP HeH motifSuperfamily: LEM domainFamily: LEM domain
14 c5k9xA_
21.7
18
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: crystal structure of tryptophan synthase alpha chain from legionella2 pneumophila subsp. pneumophila
15 c5lvaA_
21.2
10
PDB header: oxidoreductaseChain: A: PDB Molecule: nad(p)h-fmn oxidoreductase;PDBTitle: crystal structure of thermophilic tryptophan halogenase (th-hal)2 enzyme from streptomycin violaceusniger.
16 d1rtta_
20.3
11
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: NADPH-dependent FMN reductase
17 c5d88A_
19.4
23
PDB header: hydrolaseChain: A: PDB Molecule: predicted protease of the collagenase family;PDBTitle: the structure of the u32 peptidase mk0906
18 d1ni9a_
18.8
32
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: GlpX-like bacterial fructose-1,6-bisphosphatase
19 c4g6uA_
18.2
4
PDB header: toxinChain: A: PDB Molecule: ec869 cdia-ct;PDBTitle: cdia-ct/cdii toxin and immunity complex from escherichia coli
20 c4ar0A_
18.1
17
PDB header: transportChain: A: PDB Molecule: type iv pilus biogenesis and competence protein pilq;PDBTitle: n0 domain of neisseria meningitidis pilus assembly protein pilq
21 c5ohdB_
not modelled
17.8
22
PDB header: membrane proteinChain: B: PDB Molecule: growth hormone receptor;PDBTitle: putative inactive (dormant) dimeric state of ghr transmembrane domain
22 c6jdkA_
not modelled
17.6
17
PDB header: oxidoreductaseChain: A: PDB Molecule: baeyer-villiger monooxygenase;PDBTitle: crystal structure of baeyer-villiger monooxygenase from parvibaculum2 lavamentivorans
23 c4mozC_
not modelled
17.2
21
PDB header: lyaseChain: C: PDB Molecule: fructose-bisphosphate aldolase;PDBTitle: fructose-bisphosphate aldolase from slackia heliotrinireducens dsm2 20476
24 c2advB_
not modelled
16.9
33
PDB header: hydrolaseChain: B: PDB Molecule: glutaryl 7- aminocephalosporanic acid acylase;PDBTitle: crystal structures of glutaryl 7-aminocephalosporanic acid acylase:2 mutational study of activation mechanism
25 c6an7D_
not modelled
16.3
16
PDB header: transport proteinChain: D: PDB Molecule: transport permease protein;PDBTitle: crystal structure of o-antigen polysaccharide abc-transporter
26 d1h9ea_
not modelled
16.3
25
Fold: LEM/SAP HeH motifSuperfamily: LEM domainFamily: LEM domain
27 c5ltwK_
not modelled
16.1
35
PDB header: protein bindingChain: K: PDB Molecule: heat shock protein beta-6;PDBTitle: complex of human 14-3-3 sigma clu1 mutant with phosphorylated heat2 shock protein b6
28 c2outA_
not modelled
15.0
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: mu-like prophage flumu protein gp35, proteinPDBTitle: solution structure of hi1506, a novel two domain protein2 from haemophilus influenzae
29 c3f2vA_
not modelled
14.7
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: general stress protein 14;PDBTitle: crystal structure of the general stress protein 14 (tde0354) in2 complex with fmn from treponema denticola, northeast structural3 genomics consortium target tdr58.
30 c5b04G_
not modelled
14.4
19
PDB header: translationChain: G: PDB Molecule: probable translation initiation factor eif-2b subunitPDBTitle: crystal structure of the eukaryotic translation initiation factor 2b2 from schizosaccharomyces pombe
31 c6cfwI_
not modelled
13.8
23
PDB header: membrane proteinChain: I: PDB Molecule: mbh subunit;PDBTitle: cryoem structure of a respiratory membrane-bound hydrogenase
32 c3u7rB_
not modelled
13.5
17
PDB header: oxidoreductaseChain: B: PDB Molecule: nadph-dependent fmn reductase;PDBTitle: ferb - flavoenzyme nad(p)h:(acceptor) oxidoreductase (ferb) from2 paracoccus denitrificans
33 c3q9qB_
not modelled
13.2
30
PDB header: chaperoneChain: B: PDB Molecule: heat shock protein beta-1;PDBTitle: hspb1 fragment second crystal form
34 c2kncA_
not modelled
13.2
19
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
35 c6f2rT_
not modelled
13.0
20
PDB header: chaperoneChain: T: PDB Molecule: heat shock protein beta-3,heat shock protein beta-3,heatPDBTitle: a hetrotetramer of human hspb2 and hspb3
36 c5b04B_
not modelled
12.8
13
PDB header: translationChain: B: PDB Molecule: translation initiation factor eif-2b subunit alpha;PDBTitle: crystal structure of the eukaryotic translation initiation factor 2b2 from schizosaccharomyces pombe
37 c3ha2B_
not modelled
12.6
13
PDB header: oxidoreductaseChain: B: PDB Molecule: nadph-quinone reductase;PDBTitle: crystal structure of protein (nadph-quinone reductase) from2 p.pentosaceus, northeast structural genomics consortium target ptr24a
38 c2wj7D_
not modelled
12.3
35
PDB header: chaperoneChain: D: PDB Molecule: alpha-crystallin b chain;PDBTitle: human alphab crystallin
39 c5j7xA_
not modelled
12.0
17
PDB header: oxidoreductaseChain: A: PDB Molecule: dimethylaniline monooxygenase, putative;PDBTitle: baeyer-villiger monooxygenase bvmoafl838 from aspergillus flavus
40 c5mq6A_
not modelled
11.5
13
PDB header: oxidoreductaseChain: A: PDB Molecule: pyridine nucleotide-disulfide oxidoreductase-like protein;PDBTitle: polycyclic ketone monooxygenase from the thermophilic fungus2 thermothelomyces thermophila
41 c2bolA_
not modelled
11.0
26
PDB header: heat shock proteinChain: A: PDB Molecule: small heat shock protein;PDBTitle: crystal structure and assembly of tsp36, a metazoan small heat shock2 protein
42 c3a11D_
not modelled
10.8
22
PDB header: isomeraseChain: D: PDB Molecule: translation initiation factor eif-2b, delta subunit;PDBTitle: crystal structure of ribose-1,5-bisphosphate isomerase from2 thermococcus kodakaraensis kod1
43 c5m0zA_
not modelled
10.7
17
PDB header: oxidoreductaseChain: A: PDB Molecule: cyclohexanone monooxygenase from thermocrispum municipale.;PDBTitle: cyclohexanone monooxygenase from t. municipale: reduced enzyme bound2 to nadp+
44 c6daqA_
not modelled
10.3
22
PDB header: lyaseChain: A: PDB Molecule: phdj;PDBTitle: phdj bound to substrate intermediate
45 c1u0kA_
not modelled
10.2
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: gene product pa4716;PDBTitle: the structure of a predicted epimerase pa4716 from pseudomonas2 aeruginosa
46 c6c14A_
not modelled
10.2
7
PDB header: membrane protein, metal transportChain: A: PDB Molecule: protocadherin-15;PDBTitle: cryoem structure of mouse pcdh15-1ec-lhfpl5 complex
47 c1gjjA_
not modelled
9.9
25
PDB header: membrane proteinChain: A: PDB Molecule: lap2;PDBTitle: n-terminal constant region of the nuclear envelope protein2 lap2
48 c2lt2A_
not modelled
9.8
12
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: nmr structure of ba42 protein from the psychrophilic bacteria bizionia2 argentinensis sp. nov.
49 c2y9kG_
not modelled
9.7
11
PDB header: protein transportChain: G: PDB Molecule: protein invg;PDBTitle: three-dimensional model of salmonella's needle complex at subnanometer2 resolution
50 c4n6hA_
not modelled
9.7
10
PDB header: signaling proteinChain: A: PDB Molecule: soluble cytochrome b562, delta-type opioid receptorPDBTitle: 1.8 a structure of the human delta opioid 7tm receptor (psi community2 target)
51 c5tchG_
not modelled
9.6
11
PDB header: lyaseChain: G: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: crystal structure of tryptophan synthase from m. tuberculosis -2 ligand-free form, trpa-g66v mutant
52 c4b9zA_
not modelled
9.5
23
PDB header: hydrolaseChain: A: PDB Molecule: alpha-glucosidase, putative, adg31b;PDBTitle: crystal structure of agd31b, alpha-transglucosylase,2 complexed with acarbose
53 c2ks1B_
not modelled
9.4
21
PDB header: transferaseChain: B: PDB Molecule: epidermal growth factor receptor;PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
54 d1xuba1
not modelled
9.2
22
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like
55 d3bpda1
not modelled
9.2
17
Fold: Ferredoxin-likeSuperfamily: MTH889-likeFamily: MTH889-like
56 c1spfA_
not modelled
9.2
35
PDB header: lipoprotein(surface film)Chain: A: PDB Molecule: pulmonary surfactant-associated polypeptide c;PDBTitle: the nmr structure of the pulmonary surfactant-associated2 polypeptide sp-c in an apolar solvent contains a valyl-3 rich alpha-helix
57 c3vsjA_
not modelled
9.2
19
PDB header: oxidoreductaseChain: A: PDB Molecule: 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit;PDBTitle: crystal structure of 1,6-apd (2-animophenol-1,6-dioxygenase) complexed2 with intermediate products
58 c1w4xA_
not modelled
9.1
9
PDB header: oxygenaseChain: A: PDB Molecule: phenylacetone monooxygenase;PDBTitle: phenylacetone monooxygenase, a baeyer-villiger2 monooxygenase
59 d1w4xa2
not modelled
9.0
9
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains
60 c3uclA_
not modelled
8.9
26
PDB header: oxidoreductaseChain: A: PDB Molecule: cyclohexanone monooxygenase;PDBTitle: cyclohexanone-bound crystal structure of cyclohexanone monooxygenase2 in the rotated conformation
61 c6an7C_
not modelled
8.7
14
PDB header: transport proteinChain: C: PDB Molecule: transport permease protein;PDBTitle: crystal structure of o-antigen polysaccharide abc-transporter
62 c2kncB_
not modelled
8.5
26
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
63 d2raqa1
not modelled
8.5
17
Fold: Ferredoxin-likeSuperfamily: MTH889-likeFamily: MTH889-like
64 c2e7zA_
not modelled
8.4
18
PDB header: lyaseChain: A: PDB Molecule: acetylene hydratase ahy;PDBTitle: acetylene hydratase from pelobacter acetylenicus
65 c6dv5T_
not modelled
8.3
30
PDB header: chaperoneChain: T: PDB Molecule: heat shock protein beta-1;PDBTitle: oligomeric complex of a hsp27 24-mer at 3.6 a resolution
66 c5zazA_
not modelled
8.3
26
PDB header: cell adhesionChain: A: PDB Molecule: integrin beta-2;PDBTitle: solution structure of integrin b2 monomer tranmembrane domain in2 bicelle
67 c4zeoH_
not modelled
8.1
17
PDB header: translationChain: H: PDB Molecule: translation initiation factor eif-2b-like protein;PDBTitle: crystal structure of eif2b delta from chaetomium thermophilum
68 c2n2aA_
not modelled
8.1
17
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: spatial structure of her2/erbb2 dimeric transmembrane domain in the2 presence of cytoplasmic juxtamembrane domains
69 c3uoyB_
not modelled
8.0
17
PDB header: oxidoreductaseChain: B: PDB Molecule: otemo;PDBTitle: crystal structure of otemo complex with fad and nadp (form 1)
70 c3gwdA_
not modelled
8.0
26
PDB header: oxidoreductaseChain: A: PDB Molecule: cyclohexanone monooxygenase;PDBTitle: closed crystal structure of cyclohexanone monooxygenase
71 c6f2rA_
not modelled
7.9
37
PDB header: chaperoneChain: A: PDB Molecule: hspb2,heat shock protein beta-2,heat shock protein beta-2,PDBTitle: a hetrotetramer of human hspb2 and hspb3
72 c2d0gA_
not modelled
7.9
12
PDB header: hydrolaseChain: A: PDB Molecule: alpha-amylase i;PDBTitle: crystal structure of thermoactinomyces vulgaris r-47 alpha-2 amylase 1 (tvai) mutant d356n/e396q complexed with p5, a3 pullulan model oligosaccharide
73 d1pdoa_
not modelled
7.9
14
Fold: PTS system fructose IIA component-likeSuperfamily: PTS system fructose IIA component-likeFamily: EIIA-man component-like
74 c6ezoH_
not modelled
7.8
21
PDB header: membrane proteinChain: H: PDB Molecule: translation initiation factor eif-2b subunit delta;PDBTitle: eukaryotic initiation factor eif2b in complex with isrib
75 c2k74A_
not modelled
7.7
14
PDB header: membrane protein, oxidoreductaseChain: A: PDB Molecule: disulfide bond formation protein b;PDBTitle: solution nmr structure of dsbb-ubiquinone complex
76 c3fpvC_
not modelled
7.7
22
PDB header: heme binding proteinChain: C: PDB Molecule: extracellular haem-binding protein;PDBTitle: crystal structure of hbps
77 c2bbjB_
not modelled
7.7
15
PDB header: metal transport/membrane proteinChain: B: PDB Molecule: divalent cation transport-related protein;PDBTitle: crystal structure of the cora mg2+ transporter
78 c6bwsA_
not modelled
7.6
17
PDB header: unknown functionChain: A: PDB Molecule: glycolate utilization protein;PDBTitle: crystal structure of efga from methylobacterium extorquens
79 d2gm3a1
not modelled
7.3
16
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: Universal stress protein-like
80 c3gxxB_
not modelled
7.3
8
PDB header: transcriptionChain: B: PDB Molecule: transcription elongation factor spt6;PDBTitle: structure of the sh2 domain of the candida glabrata2 transcription elongation factor spt6, crystal form b
81 c2kluA_
not modelled
7.3
13
PDB header: immune system, membrane proteinChain: A: PDB Molecule: t-cell surface glycoprotein cd4;PDBTitle: nmr structure of the transmembrane and cytoplasmic domains2 of human cd4
82 c4ap3A_
not modelled
7.2
17
PDB header: oxidoreductaseChain: A: PDB Molecule: steroid monooxygenase;PDBTitle: oxidized steroid monooxygenase bound to nadp
83 c2klrA_
not modelled
7.1
37
PDB header: chaperoneChain: A: PDB Molecule: alpha-crystallin b chain;PDBTitle: solid-state nmr structure of the alpha-crystallin domain in alphab-2 crystallin oligomers
84 d1ydga_
not modelled
7.1
15
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: WrbA-like
85 c2k9yA_
not modelled
7.0
17
PDB header: transferaseChain: A: PDB Molecule: ephrin type-a receptor 2;PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
86 c2k9yB_
not modelled
7.0
17
PDB header: transferaseChain: B: PDB Molecule: ephrin type-a receptor 2;PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
87 c2z2uA_
not modelled
7.0
21
PDB header: metal binding proteinChain: A: PDB Molecule: upf0026 protein mj0257;PDBTitle: crystal structure of archaeal tyw1
88 c5ey5A_
not modelled
6.9
18
PDB header: lyaseChain: A: PDB Molecule: lbcats-a;PDBTitle: lbcats
89 c2fpgA_
not modelled
6.9
18
PDB header: ligaseChain: A: PDB Molecule: succinyl-coa ligase [gdp-forming] alpha-chain,PDBTitle: crystal structure of pig gtp-specific succinyl-coa synthetase in2 complex with gdp
90 c3ju7B_
not modelled
6.7
10
PDB header: transferaseChain: B: PDB Molecule: putative plp-dependent aminotransferase;PDBTitle: crystal structure of putative plp-dependent aminotransferase2 (np_978343.1) from bacillus cereus atcc 10987 at 2.19 a resolution
91 c6f2rE_
not modelled
6.7
37
PDB header: chaperoneChain: E: PDB Molecule: hspb2,heat shock protein beta-2,heat shock protein beta-2,PDBTitle: a hetrotetramer of human hspb2 and hspb3
92 c6f2rI_
not modelled
6.7
37
PDB header: chaperoneChain: I: PDB Molecule: hspb2,heat shock protein beta-2,heat shock protein beta-2,PDBTitle: a hetrotetramer of human hspb2 and hspb3
93 c3ptjA_
not modelled
6.6
10
PDB header: hydrolaseChain: A: PDB Molecule: upf0603 protein at1g54780, chloroplastic;PDBTitle: structural and functional analysis of arabidopsis thaliana thylakoid2 lumen protein attlp18.3
94 d1f61a_
not modelled
6.6
19
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Phosphoenolpyruvate mutase/Isocitrate lyase-like
95 c3qphA_
not modelled
6.4
11
PDB header: transcriptionChain: A: PDB Molecule: trmb, a global transcription regulator;PDBTitle: the three-dimensional structure of trmb, a global transcriptional2 regulator of the hyperthermophilic archaeon pyrococcus furiosus in3 complex with sucrose
96 c5d6aA_
not modelled
6.4
20
PDB header: hydrolaseChain: A: PDB Molecule: predicted atpase of the abc class;PDBTitle: 2.7 angstrom crystal structure of abc transporter atpase from vibrio2 vulnificus in complex with adenylyl-imidodiphosphate (amp-pnp)
97 c2l8sA_
not modelled
6.4
13
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-1;PDBTitle: solution nmr structure of transmembrane and cytosolic regions of2 integrin alpha1 in detergent micelles
98 c3e96B_
not modelled
6.1
9
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from bacillus2 clausii
99 d2j8xa1
not modelled
6.0
31
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Uracil-DNA glycosylase