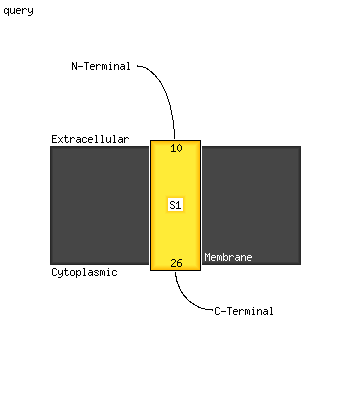
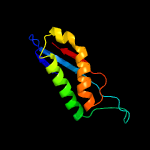
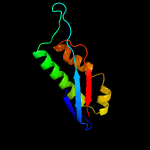
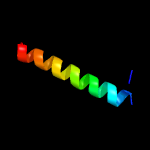
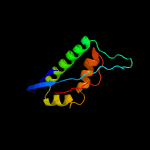
1 c3bk6C_
99.9
39
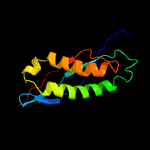
PDB header: membrane proteinChain: C: PDB Molecule: ph stomatin;PDBTitle: crystal structure of a core domain of stomatin from2 pyrococcus horikoshii
2 d1wina_
99.7
20
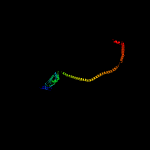
Fold: EF-Ts domain-likeSuperfamily: Band 7/SPFH domainFamily: Band 7/SPFH domain
3 c4fvjB_
99.7
38
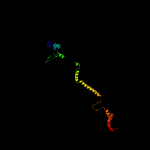
PDB header: membrane proteinChain: B: PDB Molecule: stomatin;PDBTitle: spfh domain of the mouse stomatin (crystal form 2)
4 c2rpbA_
99.7
48
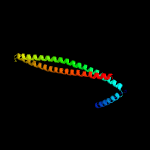
PDB header: membrane proteinChain: A: PDB Molecule: hypothetical membrane protein;PDBTitle: the solution structure of membrane protein
5 c2zv4O_
99.2
13
PDB header: structural proteinChain: O: PDB Molecule: major vault protein;PDBTitle: the structure of rat liver vault at 3.5 angstrom resolution
6 c2qzvB_
96.6
15
PDB header: structural proteinChain: B: PDB Molecule: major vault protein;PDBTitle: draft crystal structure of the vault shell at 9 angstroms2 resolution
7 c5ew5C_
75.3
13
PDB header: hydrolaseChain: C: PDB Molecule: colicin-e9;PDBTitle: crystal structure of colicin e9 in complex with its immunity protein2 im9
8 c2jp3A_
67.0
29
PDB header: transcriptionChain: A: PDB Molecule: fxyd domain-containing ion transport regulator 4;PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
9 c2jo1A_
66.7
33
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
10 c3zey0_
66.6
13
PDB header: ribosomeChain: 0: PDB Molecule: 40s ribosomal protein s3a, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
11 c2mkvA_
62.3
19
PDB header: transport proteinChain: A: PDB Molecule: sodium/potassium-transporting atpase subunit gamma;PDBTitle: structure of the na,k-atpase regulatory protein fxyd2b in micelles
12 c5xyiB_
59.2
14
PDB header: ribosomeChain: B: PDB Molecule: ribosomal protein s3ae, putative;PDBTitle: small subunit of trichomonas vaginalis ribosome
13 c3j3aB_
56.9
13
PDB header: ribosomeChain: B: PDB Molecule: 40s ribosomal protein s3a;PDBTitle: structure of the human 40s ribosomal proteins
14 c2zxeG_
55.0
38
PDB header: hydrolase/transport proteinChain: G: PDB Molecule: phospholemman-like protein;PDBTitle: crystal structure of the sodium - potassium pump in the e2.2k+.pi2 state
15 c5naoA_
51.2
22
PDB header: signaling proteinChain: A: PDB Molecule: toll-like receptor 4;PDBTitle: nmr structure of tlr4 transmembrane domain (624-657) in dpc micelles
16 c3u5gB_
44.5
22
PDB header: ribosomeChain: B: PDB Molecule: 40s ribosomal protein s1-a;PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution. this2 entry contains proteins of the 40s subunit, ribosome b
17 c5namA_
43.8
22
PDB header: signaling proteinChain: A: PDB Molecule: toll-like receptor 4;PDBTitle: nmr structure of tlr4 transmembrane domain (624-670) in dmpg/dhpc2 bicelles
18 c2xzm4_
39.9
11
PDB header: ribosomeChain: 4: PDB Molecule: 40s ribosomal protein s3a;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
19 c2lp1A_
37.3
30
PDB header: membrane proteinChain: A: PDB Molecule: c99;PDBTitle: the solution nmr structure of the transmembrane c-terminal domain of2 the amyloid precursor protein (c99)
20 c3n23E_
34.8
30
PDB header: hydrolaseChain: E: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the high affinity complex between ouabain and the2 e2p form of the sodium-potassium pump
21 c2n28A_
not modelled
34.3
31
PDB header: viral proteinChain: A: PDB Molecule: protein vpu;PDBTitle: solid-state nmr structure of vpu
22 c3kdpD_
not modelled
33.8
14
PDB header: hydrolaseChain: D: PDB Molecule: sodium/potassium-transporting atpase subunit beta-1;PDBTitle: crystal structure of the sodium-potassium pump
23 c6mq2D_
not modelled
33.2
33
PDB header: de novo proteinChain: D: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-2
24 c6mctC_
not modelled
33.2
33
PDB header: de novo proteinChain: C: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
25 c6mctD_
not modelled
33.2
33
PDB header: de novo proteinChain: D: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
26 c6mctE_
not modelled
33.2
33
PDB header: de novo proteinChain: E: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
27 c6mctB_
not modelled
33.2
33
PDB header: de novo proteinChain: B: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
28 c6mctF_
not modelled
33.2
33
PDB header: de novo proteinChain: F: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
29 c6mctG_
not modelled
33.2
33
PDB header: de novo proteinChain: G: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
30 c6mctJ_
not modelled
33.2
33
PDB header: de novo proteinChain: J: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
31 c6mctL_
not modelled
33.2
33
PDB header: de novo proteinChain: L: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
32 c6mctI_
not modelled
33.2
33
PDB header: de novo proteinChain: I: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
33 c6mctO_
not modelled
33.2
33
PDB header: de novo proteinChain: O: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
34 c6mctH_
not modelled
33.2
33
PDB header: de novo proteinChain: H: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
35 c6mpwA_
not modelled
33.2
33
PDB header: de novo proteinChain: A: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-1
36 c6mctK_
not modelled
33.2
33
PDB header: de novo proteinChain: K: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
37 c6mctM_
not modelled
33.2
33
PDB header: de novo proteinChain: M: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
38 c6mctN_
not modelled
33.2
33
PDB header: de novo proteinChain: N: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
39 c6mctA_
not modelled
33.2
33
PDB header: de novo proteinChain: A: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
40 c6ahqM_
not modelled
31.8
13
PDB header: motor proteinChain: M: PDB Molecule: flagellar protein flil;PDBTitle: structure of the 40-167 fragment of flil
41 d2c1ia2
not modelled
31.8
19
Fold: Peptidoglycan deacetylase N-terminal noncatalytic regionSuperfamily: Peptidoglycan deacetylase N-terminal noncatalytic regionFamily: Peptidoglycan deacetylase N-terminal noncatalytic region
42 c2lz3B_
not modelled
31.0
30
PDB header: membrane proteinChain: B: PDB Molecule: amyloid beta a4 protein;PDBTitle: solution nmr structure of transmembrane domain of amyloid precursor2 protein wt
43 c2lz3A_
not modelled
31.0
30
PDB header: membrane proteinChain: A: PDB Molecule: amyloid beta a4 protein;PDBTitle: solution nmr structure of transmembrane domain of amyloid precursor2 protein wt
44 c4pj0L_
not modelled
29.5
21
PDB header: oxidoreductase, electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: structure of t.elongatus photosystem ii, rows of dimers crystal2 packing
45 c4pj0l_
not modelled
29.5
21
PDB header: oxidoreductase, electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: structure of t.elongatus photosystem ii, rows of dimers crystal2 packing
46 c2lz4B_
not modelled
29.1
30
PDB header: membrane proteinChain: B: PDB Molecule: amyloid beta a4 protein;PDBTitle: solution nmr structure of transmembrane domain of amyloid precursor2 protein v44m
47 c2lz4A_
not modelled
29.1
30
PDB header: membrane proteinChain: A: PDB Molecule: amyloid beta a4 protein;PDBTitle: solution nmr structure of transmembrane domain of amyloid precursor2 protein v44m
48 c3wu2L_
not modelled
28.8
21
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure analysis of photosystem ii complex
49 c4ub8l_
not modelled
28.8
21
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: native structure of photosystem ii (dataset-2) by a femtosecond x-ray2 laser
50 c3a0hL_
not modelled
28.8
21
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of i-substituted photosystem ii complex
51 c5e7cL_
not modelled
28.8
21
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: macromolecular diffractive imaging using imperfect crystals - bragg2 data
52 c4rvyL_
not modelled
28.8
21
PDB header: oxidoreductaseChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: serial time resolved crystallography of photosystem ii using a2 femtosecond x-ray laser. the s state after two flashes (s3)
53 c5e7cl_
not modelled
28.8
21
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: macromolecular diffractive imaging using imperfect crystals - bragg2 data
54 c4ixrL_
not modelled
28.8
21
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt fs x-ray diffraction of photosystem ii, first illuminated state
55 c4ub8L_
not modelled
28.8
21
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: native structure of photosystem ii (dataset-2) by a femtosecond x-ray2 laser
56 c3a0bL_
not modelled
28.8
21
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of br-substituted photosystem ii complex
57 c3a0hl_
not modelled
28.8
21
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of i-substituted photosystem ii complex
58 c4rvyl_
not modelled
28.8
21
PDB header: oxidoreductaseChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: serial time resolved crystallography of photosystem ii using a2 femtosecond x-ray laser. the s state after two flashes (s3)
59 c3prqL_
not modelled
28.8
21
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 1 of 2). this file contains first monomer of psii3 dimer
60 d2axtl1
not modelled
28.8
21
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein L, PsbLFamily: PsbL-like
61 c3wu2l_
not modelled
28.8
21
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure analysis of photosystem ii complex
62 c4ub6L_
not modelled
28.8
21
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: native structure of photosystem ii (dataset-1) by a femtosecond x-ray2 laser
63 c3bz2L_
not modelled
28.8
21
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 2 of 2). this2 file contains second monomer of psii dimer
64 c4fbyd_
not modelled
28.8
21
PDB header: photosynthesisChain: D: PDB Molecule: photosystem ii d2 protein;PDBTitle: fs x-ray diffraction of photosystem ii
65 c4tnjl_
not modelled
28.8
21
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii 500 ms after the 2nd illumination2 (2f) at 4.5 a resolution
66 c3arcL_
not modelled
28.8
21
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
67 c2axtL_
not modelled
28.8
21
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
68 c3kziL_
not modelled
28.8
21
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of monomeric form of cyanobacterial photosystem ii
69 c2axtl_
not modelled
28.8
21
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
70 c1s5lL_
not modelled
28.8
21
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
71 c4ub6l_
not modelled
28.8
21
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: native structure of photosystem ii (dataset-1) by a femtosecond x-ray2 laser
72 c4tnkL_
not modelled
28.8
21
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii 250 microsec after the third2 illumination at 5.2 a resolution
73 c4tnhl_
not modelled
28.8
21
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii in the dark state at 4.9 a2 resolution
74 c4tniL_
not modelled
28.8
21
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii 500 ms after the third2 illumination at 4.6 a resolution
75 c4tnhL_
not modelled
28.8
21
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii in the dark state at 4.9 a2 resolution
76 c4fbyL_
not modelled
28.8
21
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: fs x-ray diffraction of photosystem ii
77 c4tnkl_
not modelled
28.8
21
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii 250 microsec after the third2 illumination at 5.2 a resolution
78 c4tnil_
not modelled
28.8
21
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii 500 ms after the third2 illumination at 4.6 a resolution
79 c4tnjL_
not modelled
28.8
21
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii 500 ms after the 2nd illumination2 (2f) at 4.5 a resolution
80 c1s5ll_
not modelled
28.8
21
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
81 c4il6L_
not modelled
28.8
21
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: structure of sr-substituted photosystem ii
82 c3bz1L_
not modelled
28.8
21
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 1 of 2). this2 file contains first monomer of psii dimer
83 c3a0bl_
not modelled
28.8
21
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of br-substituted photosystem ii complex
84 c3prrL_
not modelled
28.8
21
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 2 of 2). this file contains second monomer of psii3 dimer
85 c4ixql_
not modelled
28.8
21
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt fs x-ray diffraction of photosystem ii, dark state
86 c4ixqL_
not modelled
28.8
21
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt fs x-ray diffraction of photosystem ii, dark state
87 c4ixrl_
not modelled
28.8
21
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt fs x-ray diffraction of photosystem ii, first illuminated state
88 c4il6l_
not modelled
28.8
21
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: structure of sr-substituted photosystem ii
89 c2lohA_
not modelled
27.1
30
PDB header: neuropeptideChain: A: PDB Molecule: p3(42);PDBTitle: dimeric structure of transmembrane domain of amyloid precursor protein2 in micellar environment
90 c6nhwE_
not modelled
26.7
15
PDB header: immune systemChain: E: PDB Molecule: tumor necrosis factor receptor superfamily member 10b;PDBTitle: structure of the transmembrane domain of the death receptor 5 - dimer2 of trimer
91 c2lx0A_
not modelled
25.5
33
PDB header: membrane proteinChain: A: PDB Molecule: membrane fusion protein p14;PDBTitle: arced helix (arch) nmr structure of the reovirus p14 fusion-associated2 small transmembrane (fast) protein transmembrane domain (tmd) in3 dodecyl phosphocholine (dpc) micelles
92 c2m59A_
not modelled
25.3
11
PDB header: transferaseChain: A: PDB Molecule: vascular endothelial growth factor receptor 2;PDBTitle: spatial structure of dimeric vegfr2 membrane domain in dpc micelles
93 c2m59B_
not modelled
25.3
11
PDB header: transferaseChain: B: PDB Molecule: vascular endothelial growth factor receptor 2;PDBTitle: spatial structure of dimeric vegfr2 membrane domain in dpc micelles
94 c6nhwA_
not modelled
25.1
29
PDB header: immune systemChain: A: PDB Molecule: tumor necrosis factor receptor superfamily member 10b;PDBTitle: structure of the transmembrane domain of the death receptor 5 - dimer2 of trimer
95 c6nhwB_
not modelled
25.1
29
PDB header: immune systemChain: B: PDB Molecule: tumor necrosis factor receptor superfamily member 10b;PDBTitle: structure of the transmembrane domain of the death receptor 5 - dimer2 of trimer
96 c6nhwC_
not modelled
25.1
29
PDB header: immune systemChain: C: PDB Molecule: tumor necrosis factor receptor superfamily member 10b;PDBTitle: structure of the transmembrane domain of the death receptor 5 - dimer2 of trimer
97 c6hu9e_
not modelled
24.9
14
PDB header: oxidoreductase/electron transportChain: E: PDB Molecule: cytochrome b-c1 complex subunit rieske, mitochondrial;PDBTitle: iii2-iv2 mitochondrial respiratory supercomplex from s. cerevisiae
98 c6g52H_
not modelled
23.3
14
PDB header: metal transportChain: H: PDB Molecule: metal transporter cnnm4;PDBTitle: crystal structure of the cnmp binding domain of the magnesium2 transporter cnnm4
99 c6iycE_
not modelled
22.9
30
PDB header: membrane proteinChain: E: PDB Molecule: amyloid-beta a4 protein;PDBTitle: recognition of the amyloid precursor protein by human gamma-secretase
100 c3arcl_
not modelled
22.9
21
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
101 c3j00Z_
not modelled
22.1
17
PDB header: ribosome/ribosomal proteinChain: Z: PDB Molecule: cell division protein ftsq;PDBTitle: structure of the ribosome-secye complex in the membrane environment
102 c6nhwD_
not modelled
22.1
15
PDB header: immune systemChain: D: PDB Molecule: tumor necrosis factor receptor superfamily member 10b;PDBTitle: structure of the transmembrane domain of the death receptor 5 - dimer2 of trimer
103 c6nhwF_
not modelled
22.1
15
PDB header: immune systemChain: F: PDB Molecule: tumor necrosis factor receptor superfamily member 10b;PDBTitle: structure of the transmembrane domain of the death receptor 5 - dimer2 of trimer
104 c3kdpH_
not modelled
21.8
32
PDB header: hydrolaseChain: H: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the sodium-potassium pump
105 c3kdpG_
not modelled
21.8
32
PDB header: hydrolaseChain: G: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the sodium-potassium pump
106 c3jcul_
not modelled
21.8
21
PDB header: membrane proteinChain: L: PDB Molecule: protein photosystem ii reaction center protein l;PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
107 d3ecfa1
not modelled
21.6
17
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Ava4193-like
108 d2f1la1
not modelled
21.4
17
Fold: PRC-barrel domainSuperfamily: PRC-barrel domainFamily: RimM C-terminal domain-like
109 c6nhyC_
not modelled
21.3
15
PDB header: immune systemChain: C: PDB Molecule: tumor necrosis factor receptor superfamily member 10b;PDBTitle: structure of the transmembrane domain of the death receptor 5 mutant2 (g217y) - trimer only
110 c6nhyA_
not modelled
21.3
15
PDB header: immune systemChain: A: PDB Molecule: tumor necrosis factor receptor superfamily member 10b;PDBTitle: structure of the transmembrane domain of the death receptor 5 mutant2 (g217y) - trimer only
111 c6nhyB_
not modelled
21.3
15
PDB header: immune systemChain: B: PDB Molecule: tumor necrosis factor receptor superfamily member 10b;PDBTitle: structure of the transmembrane domain of the death receptor 5 mutant2 (g217y) - trimer only
112 c3j20A_
not modelled
21.1
7
PDB header: ribosomeChain: A: PDB Molecule: 30s ribosomal protein s3ae;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (30s3 ribosomal subunit)
113 c6mpwE_
not modelled
20.9
33
PDB header: de novo proteinChain: E: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-1
114 c6mq2A_
not modelled
20.9
33
PDB header: de novo proteinChain: A: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-2
115 c6mpwD_
not modelled
20.9
33
PDB header: de novo proteinChain: D: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-1
116 c6mq2C_
not modelled
20.9
33
PDB header: de novo proteinChain: C: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-2
117 c6mpwC_
not modelled
20.9
33
PDB header: de novo proteinChain: C: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-1
118 c6mq2B_
not modelled
20.9
33
PDB header: de novo proteinChain: B: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-2
119 c6mpwB_
not modelled
20.9
33
PDB header: de novo proteinChain: B: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-1
120 c6mq2E_
not modelled
20.9
33
PDB header: de novo proteinChain: E: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-2