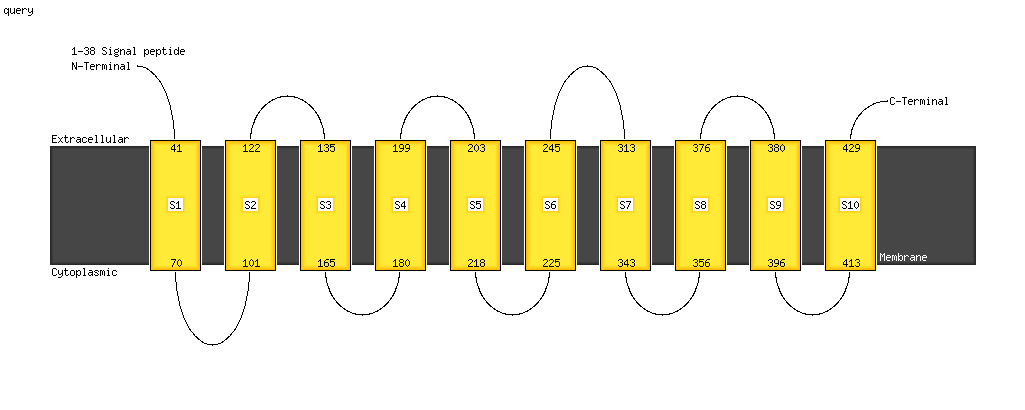
1 c4kppA_
41.6
10
PDB header: membrane proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of h+/ca2+ exchanger cax
2 c6iu3A_
37.4
16
PDB header: metal transportChain: A: PDB Molecule: vit1;PDBTitle: crystal structure of iron transporter vit1 with zinc ions
3 c5jyfB_
33.1
22
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: structures of streptococcus agalactiae gbs gapdh in different2 enzymatic states
4 c1rm4O_
32.8
26
PDB header: oxidoreductaseChain: O: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase a;PDBTitle: crystal structure of recombinant photosynthetic glyceraldehyde-3-2 phosphate dehydrogenase a4 isoform, complexed with nadp
5 c3hq4R_
32.5
26
PDB header: oxidoreductaseChain: R: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase 1;PDBTitle: crystal structure of c151s mutant of glyceraldehyde-3-phosphate2 dehydrogenase 1 (gapdh1) complexed with nad from staphylococcus3 aureus mrsa252 at 2.2 angstrom resolution
6 c2pkrI_
32.0
26
PDB header: oxidoreductaseChain: I: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase aor;PDBTitle: crystal structure of (a+cte)4 chimeric form of photosyntetic2 glyceraldehyde-3-phosphate dehydrogenase, complexed with nadp
7 c4qx6A_
31.2
22
PDB header: oxidoreductaseChain: A: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 streptococcus agalactiae nem316 at 2.46 angstrom resolution
8 c5ld5C_
31.1
13
PDB header: oxidoreductaseChain: C: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of a bacterial dehydrogenase at 2.19 angstroms2 resolution
9 c2x5kO_
30.7
13
PDB header: oxidoreductaseChain: O: PDB Molecule: d-erythrose-4-phosphate dehydrogenase;PDBTitle: structure of an active site mutant of the d-erythrose-4-phosphate2 dehydrogenase from e. coli
10 c4dibF_
30.3
26
PDB header: oxidoreductaseChain: F: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: the crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 bacillus anthracis str. sterne
11 c1hdgO_
30.3
17
PDB header: oxidoreductase (aldehy(d)-nad(a))Chain: O: PDB Molecule: holo-d-glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: the crystal structure of holo-glyceraldehyde-3-phosphate dehydrogenase2 from the hyperthermophilic bacterium thermotoga maritima at 2.53 angstroms resolution
12 c3cieC_
30.2
22
PDB header: oxidoreductaseChain: C: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde 3-phosphate2 dehydrogenase from cryptosporidium parvum
13 c3docD_
30.0
13
PDB header: oxidoreductaseChain: D: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: crystal structure of trka glyceraldehyde-3-phosphate dehydrogenase2 from brucella melitensis
14 c5ur0B_
29.7
22
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystallographic structure of glyceraldehyde-3-phosphate dehydrogenase2 from naegleria gruberi
15 c3b20R_
29.4
26
PDB header: oxidoreductaseChain: R: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase (nadp+);PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase2 complexed with nadfrom synechococcus elongatus"
16 c3h9eO_
29.3
13
PDB header: oxidoreductaseChain: O: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase, testis-specific;PDBTitle: crystal structure of human sperm-specific glyceraldehyde-3-phosphate2 dehydrogenase (gapds) complex with nad and phosphate
17 c2d2iO_
29.2
26
PDB header: oxidoreductaseChain: O: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: crystal structure of nadp-dependent glyceraldehyde-3-2 phosphate dehydrogenase from synechococcus sp. complexed3 with nadp+
18 d2ca1a1
29.1
20
Fold: Nucleocapsid protein dimerization domainSuperfamily: Nucleocapsid protein dimerization domainFamily: Coronavirus nucleocapsid protein
19 c1cerC_
29.0
17
PDB header: oxidoreductase (aldehyde(d)-nad(a))Chain: C: PDB Molecule: holo-d-glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: determinants of enzyme thermostability observed in the2 molecular structure of thermus aquaticus d-glyceraldehyde-3 3-phosphate dehydrogenase at 2.5 angstroms resolution
20 c2ep7B_
28.9
26
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: structural study of project id aq_1065 from aquifex aeolicus vf5
21 c2b4rQ_
not modelled
28.9
13
PDB header: oxidoreductaseChain: Q: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 plasmodium falciparum at 2.25 angstrom resolution reveals intriguing3 extra electron density in the active site
22 c1i32D_
not modelled
28.8
30
PDB header: oxidoreductaseChain: D: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: leishmania mexicana glyceraldehyde-3-phosphate2 dehydrogenase in complex with inhibitors
23 d2ge7a1
not modelled
28.7
20
Fold: Nucleocapsid protein dimerization domainSuperfamily: Nucleocapsid protein dimerization domainFamily: Coronavirus nucleocapsid protein
24 c6ok4A_
not modelled
28.7
9
PDB header: oxidoreductaseChain: A: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase (gapdh)2 from chlamydia trachomatis with bound nad
25 c1ihxD_
not modelled
28.5
13
PDB header: oxidoreductaseChain: D: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: crystal structure of two d-glyceraldehyde-3-phosphate2 dehydrogenase complexes: a case of asymmetry
26 c2i5pO_
not modelled
28.2
22
PDB header: oxidoreductaseChain: O: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase 1;PDBTitle: crystal structure of glyceraldehyde-3-phosphate2 dehydrogenase isoform 1 from k. marxianus
27 c1s7cA_
not modelled
26.5
13
PDB header: structural genomics, oxidoreductaseChain: A: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase a;PDBTitle: crystal structure of mes buffer bound form of glyceraldehyde 3-2 phosphate dehydrogenase from escherichia coli
28 d1kqfb2
not modelled
26.3
32
Fold: Single transmembrane helixSuperfamily: Iron-sulfur subunit of formate dehydrogenase N, transmembrane anchorFamily: Iron-sulfur subunit of formate dehydrogenase N, transmembrane anchor
29 c1obfO_
not modelled
24.8
17
PDB header: glycolytic pathwayChain: O: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: the crystal structure of glyceraldehyde 3-phosphate2 dehydrogenase from alcaligenes xylosoxidans at 1.7 a3 resolution.
30 c5epwB_
not modelled
22.8
36
PDB header: viral proteinChain: B: PDB Molecule: nucleoprotein;PDBTitle: c-terminal domain of human coronavirus nl63 nucleocapsid protein
31 c3hjaB_
not modelled
22.7
16
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 borrelia burgdorferi
32 c5j9gB_
not modelled
22.0
13
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-p dehydrogenase;PDBTitle: structure of lactobacillus acidophilus glyceraldehyde-3-phosphate2 dehydrogenase at 2.21 angstrom resolution
33 c3sthA_
not modelled
18.5
22
PDB header: oxidoreductaseChain: A: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 toxoplasma gondii
34 c2kr7A_
not modelled
17.6
33
PDB header: isomeraseChain: A: PDB Molecule: fkbp-type peptidyl-prolyl cis-trans isomerase slyd;PDBTitle: solution structure of helicobacter pylori slyd
35 c1r7gA_
not modelled
16.9
63
PDB header: membrane proteinChain: A: PDB Molecule: genome polyprotein;PDBTitle: nmr structure of the membrane anchor domain (1-31) of the2 nonstructural protein 5a (ns5a) of hepatitis c virus3 (minimized average structure, sample in 100mm dpc)
36 c1r7cA_
not modelled
16.9
63
PDB header: membrane proteinChain: A: PDB Molecule: genome polyprotein;PDBTitle: nmr structure of the membrane anchor domain (1-31) of the2 nonstructural protein 5a (ns5a) of hepatitis c virus3 (minimized average structure, sample in 50% tfe)
37 c2gd1P_
not modelled
16.7
23
PDB header: oxidoreductase(aldehyde(d)-nad(a))Chain: P: PDB Molecule: apo-d-glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: coenzyme-induced conformational changes in glyceraldehyde-3-2 phosphate dehydrogenase from bacillus stearothermophillus
38 c2ntxB_
not modelled
16.7
22
PDB header: signaling proteinChain: B: PDB Molecule: emb
39 c4hkrA_
not modelled
16.5
21
PDB header: transport proteinChain: A: PDB Molecule: calcium release-activated calcium channel protein 1;PDBTitle: calcium release-activated calcium (crac) channel orai
40 c4h44E_
not modelled
16.5
50
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: 2.70 a cytochrome b6f complex structure from nostoc pcc 7120
41 c2zt9E_
not modelled
16.5
50
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex from nostoc sp. pcc2 7120
42 c4ogqE_
not modelled
16.5
50
PDB header: electron transportChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: internal lipid architecture of the hetero-oligomeric cytochrome b6f2 complex
43 d2giba1
not modelled
15.4
29
Fold: Nucleocapsid protein dimerization domainSuperfamily: Nucleocapsid protein dimerization domainFamily: Coronavirus nucleocapsid protein
44 c4ci7A_
not modelled
14.5
17
PDB header: hydrolaseChain: A: PDB Molecule: cell surface protein (putative cell surface-associatedPDBTitle: the crystal structure of the cysteine protease and lectin-like2 domains of cwp84, a surface layer associated protein of clostridium3 difficile
45 c3pr9A_
not modelled
14.1
11
PDB header: chaperoneChain: A: PDB Molecule: fkbp-type peptidyl-prolyl cis-trans isomerase;PDBTitle: structural analysis of protein folding by the methanococcus jannaschii2 chaperone fkbp26
46 d2cjra1
not modelled
13.9
29
Fold: Nucleocapsid protein dimerization domainSuperfamily: Nucleocapsid protein dimerization domainFamily: Coronavirus nucleocapsid protein
47 d1pvhb_
not modelled
13.9
14
Fold: 4-helical cytokinesSuperfamily: 4-helical cytokinesFamily: Long-chain cytokines
48 d3buxb1
not modelled
13.8
36
Fold: EF Hand-likeSuperfamily: EF-handFamily: EF-hand modules in multidomain proteins
49 d2h1qa1
not modelled
13.0
26
Fold: PLP-dependent transferase-likeSuperfamily: Dhaf3308-likeFamily: Dhaf3308-like
50 c6g13B_
not modelled
12.9
26
PDB header: viral proteinChain: B: PDB Molecule: nucleoprotein;PDBTitle: c-terminal domain of mers-cov nucleocapsid
51 c4olsA_
not modelled
12.0
28
PDB header: hydrolaseChain: A: PDB Molecule: endolysin;PDBTitle: the amidase-2 domain of lysgh15
52 c5i0cA_
not modelled
11.9
16
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized protein yjdj;PDBTitle: crystal structure of predicted acyltransferase yjdj with acyl-coa n-2 acyltransferase domain from escherichia coli str. k-12
53 c6g9oA_
not modelled
11.7
28
PDB header: membrane proteinChain: A: PDB Molecule: volume-regulated anion channel subunit lrrc8a;PDBTitle: structure of full-length homomeric mlrrc8a volume-regulated anion2 channel at 4.25 a resolution
54 d2auwa1
not modelled
11.7
25
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: NE0471 C-terminal domain-like
55 c3wmjB_
not modelled
11.6
24
PDB header: viral proteinChain: B: PDB Molecule: eiav vaccine gp45;PDBTitle: crystal structure of eiav vaccine gp45
56 d1a7ja_
not modelled
10.3
23
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Phosphoribulokinase/pantothenate kinase
57 c3srjB_
not modelled
9.8
29
PDB header: cell invasion/inhibitorChain: B: PDB Molecule: apical membrane antigen 1, ama1;PDBTitle: pfama1 in complex with invasion-inhibitory peptide r1
58 c4jo6Z_
not modelled
9.5
50
PDB header: unknown functionChain: Z: PDB Molecule: sbp-tag;PDBTitle: streptavidin complex with sbp-tag
59 c4jo6Y_
not modelled
9.5
50
PDB header: unknown functionChain: Y: PDB Molecule: sbp-tag;PDBTitle: streptavidin complex with sbp-tag
60 c5n5xN_
not modelled
9.3
40
PDB header: transcriptionChain: N: PDB Molecule: rna polymerase i-specific transcription initiation factorPDBTitle: crystal structure of s. cerevisiae core factor at 3.2a resolution
61 c5yq7H_
not modelled
9.1
65
PDB header: photosynthesisChain: H: PDB Molecule: alpha subunit of light-harvesting 1;PDBTitle: cryo-em structure of the rc-lh core complex from roseiflexus2 castenholzii
62 d1cwpa_
not modelled
8.9
42
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Bromoviridae-like VP
63 c5vlaZ_
not modelled
8.9
58
PDB header: hydrolaseChain: Z: PDB Molecule: thr-val-phe-thr-ser-trp-glu-glu-tyr-leu-asp-trp-val-met-PDBTitle: short pcsk9 delta-p' complex with fusion2 peptide
64 d1r3ba_
not modelled
8.9
23
Fold: Bromodomain-likeSuperfamily: Mob1/phoceinFamily: Mob1/phocein
65 d1wjva1
not modelled
8.8
18
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: C2HC finger
66 c3npgD_
not modelled
8.8
26
PDB header: unknown functionChain: D: PDB Molecule: uncharacterized duf364 family protein;PDBTitle: crystal structure of a protein with unknown function from duf3642 family (ph1506) from pyrococcus horikoshii at 2.70 a resolution
67 c2k8iA_
not modelled
8.8
11
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase;PDBTitle: solution structure of e.coli slyd
68 d1ix5a_
not modelled
8.7
11
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase
69 d1xmta_
not modelled
8.6
15
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-acetyl transferase, NAT
70 c2kvlA_
not modelled
8.5
27
PDB header: viral proteinChain: A: PDB Molecule: major outer capsid protein vp7;PDBTitle: nmr structure of the c-terminal domain of vp7
71 c2jagA_
not modelled
8.5
12
PDB header: membrane proteinChain: A: PDB Molecule: halorhodopsin;PDBTitle: l1-intermediate of halorhodopsin t203v
72 d1wxxa1
not modelled
8.4
55
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: Hypothetical RNA methyltransferase domain (HRMD)
73 d2fmma1
not modelled
8.4
31
Fold: SH3-like barrelSuperfamily: Chromo domain-likeFamily: Chromo domain
74 c3cgnA_
not modelled
8.4
22
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase;PDBTitle: crystal structure of thermophilic slyd
75 c2kfwA_
not modelled
8.3
11
PDB header: isomeraseChain: A: PDB Molecule: fkbp-type peptidyl-prolyl cis-trans isomerasePDBTitle: solution structure of full-length slyd from e.coli
76 c6go1A_
not modelled
8.3
22
PDB header: hydrolaseChain: A: PDB Molecule: polysaccharide deacetylase-like protein;PDBTitle: crystal structure of a bacillus anthracis peptidoglycan deacetylase
77 c4ln0C_
not modelled
8.2
34
PDB header: transcriptionChain: C: PDB Molecule: transcription cofactor vestigial-like protein 4;PDBTitle: crystal structure of the vgll4-tead4 complex
78 c3prdA_
not modelled
8.2
13
PDB header: chaperone, isomeraseChain: A: PDB Molecule: fkbp-type peptidyl-prolyl cis-trans isomerase;PDBTitle: structural analysis of protein folding by the methanococcus jannaschii2 chaperone fkbp26
79 d1fhfa_
not modelled
7.8
13
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: CCP-like
80 c1cwpB_
not modelled
7.8
42
PDB header: virus/rnaChain: B: PDB Molecule: coat protein;PDBTitle: structures of the native and swollen forms of cowpea2 chlorotic mottle virus determined by x-ray crystallography3 and cryo-electron microscopy
81 c4dt4A_
not modelled
7.7
38
PDB header: isomeraseChain: A: PDB Molecule: fkbp-type 16 kda peptidyl-prolyl cis-trans isomerase;PDBTitle: crystal structure of the ppiase-chaperone slpa with the chaperone2 binding site occupied by the linker of the purification tag
82 c3ksyA_
not modelled
7.7
34
PDB header: signaling proteinChain: A: PDB Molecule: son of sevenless homolog 1;PDBTitle: crystal structure of the histone domain, dh-ph unit, and catalytic2 unit of the ras activator son of sevenless (sos)
83 c4g10A_
not modelled
7.6
20
PDB header: transferaseChain: A: PDB Molecule: glutathione s-transferase homolog;PDBTitle: ligg from sphingobium sp. syk-6 is related to the glutathione2 transferase omega class
84 c3u4gA_
not modelled
7.6
50
PDB header: transferaseChain: A: PDB Molecule: namn:dmb phosphoribosyltransferase;PDBTitle: the structure of cobt from pyrococcus horikoshii
85 d1e12a_
not modelled
7.6
11
Fold: Family A G protein-coupled receptor-likeSuperfamily: Family A G protein-coupled receptor-likeFamily: Bacteriorhodopsin-like
86 d1uj8a1
not modelled
7.5
38
Fold: Another 3-helical bundleSuperfamily: IscX-likeFamily: IscX-like
87 c4hkrB_
not modelled
7.3
21
PDB header: transport proteinChain: B: PDB Molecule: calcium release-activated calcium channel protein 1;PDBTitle: calcium release-activated calcium (crac) channel orai
88 d1yc611
not modelled
7.2
42
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Bromoviridae-like VP
89 c6e2qP_
not modelled
6.9
14
PDB header: signaling proteinChain: P: PDB Molecule: erythropoietin receptor;PDBTitle: structure of human jak2 ferm/sh2 in complex with erythropoietin2 receptor
90 c1p7bB_
not modelled
6.7
20
PDB header: metal transportChain: B: PDB Molecule: integral membrane channel and cytosolic domains;PDBTitle: crystal structure of an inward rectifier potassium channel
91 c5j8hB_
not modelled
6.5
50
PDB header: metal binding protein/transferaseChain: B: PDB Molecule: eukaryotic elongation factor 2 kinase;PDBTitle: structure of calmodulin in a complex with a peptide derived from a2 calmodulin-dependent kinase
92 c3blxM_
not modelled
6.4
22
PDB header: oxidoreductaseChain: M: PDB Molecule: isocitrate dehydrogenase [nad] subunit 1;PDBTitle: yeast isocitrate dehydrogenase (apo form)
93 d1js9c_
not modelled
6.4
42
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Bromoviridae-like VP
94 c5vf3Z_
not modelled
6.3
60
PDB header: virusChain: Z: PDB Molecule: highly immunogenic outer capsid protein;PDBTitle: bacteriophage t4 isometric capsid
95 c2mwiA_
not modelled
6.1
67
PDB header: protein bindingChain: A: PDB Molecule: deoxynucleotidyltransferase terminal-interacting protein 1;PDBTitle: the structure of the carboxy-terminal domain of dnttip1
96 d3d37a1
not modelled
6.0
18
Fold: Phage tail proteinsSuperfamily: Phage tail proteinsFamily: Baseplate protein-like
97 c6dv5T_
not modelled
5.9
17
PDB header: chaperoneChain: T: PDB Molecule: heat shock protein beta-1;PDBTitle: oligomeric complex of a hsp27 24-mer at 3.6 a resolution
98 c1w5cL_
not modelled
5.9
42
PDB header: photosynthesisChain: L: PDB Molecule: cytochrome b559 beta subunit;PDBTitle: photosystem ii from thermosynechococcus elongatus
99 c6adqP_
not modelled
5.9
27
PDB header: electron transportChain: P: PDB Molecule: prokaryotic respiratory supercomplex associate factor 1PDBTitle: respiratory complex ciii2civ2sod2 from mycobacterium smegmatis