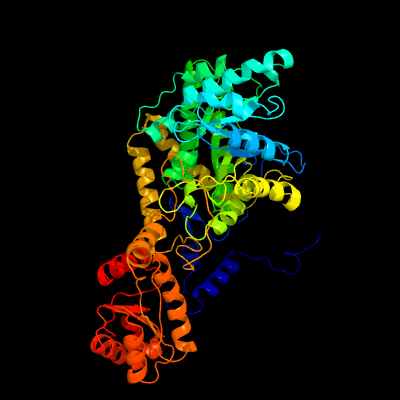
| 1 | c6reqB_
|
|
 |
100.0 |
33 |
PDB header:isomerase
Chain: B: PDB Molecule:protein (methylmalonyl-coa mutase);
PDBTitle: methylmalonyl-coa mutase, 3-carboxypropyl-coa inhibitor complex
|
|
|
|
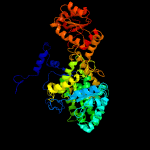
| 2 | c3bicA_
|
|
 |
100.0 |
26 |
PDB header:isomerase
Chain: A: PDB Molecule:methylmalonyl-coa mutase, mitochondrial precursor;
PDBTitle: crystal structure of human methylmalonyl-coa mutase
|
|
|
|
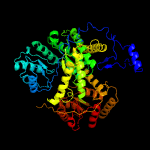
| 3 | c1e1cA_
|
|
 |
100.0 |
26 |
PDB header:isomerase
Chain: A: PDB Molecule:methylmalonyl-coa mutase alpha chain;
PDBTitle: methylmalonyl-coa mutase h244a mutant
|
|
|
|
| 4 | d7reqb1
|
|
 |
100.0 |
34 |
Fold:TIM beta/alpha-barrel
Superfamily:Cobalamin (vitamin B12)-dependent enzymes
Family:Methylmalonyl-CoA mutase, N-terminal (CoA-binding) domain |
|
|
|
| 5 | d7reqa1
|
|
 |
100.0 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:Cobalamin (vitamin B12)-dependent enzymes
Family:Methylmalonyl-CoA mutase, N-terminal (CoA-binding) domain |
|
|
|
| 6 | c4r3uB_
|
|
 |
100.0 |
26 |
PDB header:isomerase
Chain: B: PDB Molecule:2-hydroxyisobutyryl-coa mutase large subunit;
PDBTitle: crystal structure of 2-hydroxyisobutyryl-coa mutase
|
|
|
|
| 7 | c4xc8B_
|
|
 |
100.0 |
22 |
PDB header:isomerase
Chain: B: PDB Molecule:isobutyryl-coa mutase fused;
PDBTitle: isobutyryl-coa mutase fused with bound butyryl-coa, gdp, and mg and2 without cobalamin (apo-icmf/gdp)
|
|
|
|
| 8 | d7reqb2
|
|
 |
100.0 |
29 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
|
| 9 | d7reqa2
|
|
 |
100.0 |
15 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
|
| 10 | d1ccwa_
|
|
 |
99.2 |
11 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
|
| 11 | d1xrsb1
|
|
 |
98.9 |
18 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
|
| 12 | c2yxbA_
|
|
 |
98.8 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:coenzyme b12-dependent mutase;
PDBTitle: crystal structure of the methylmalonyl-coa mutase alpha-subunit from2 aeropyrum pernix
|
|
|
|
| 13 | c1xrsB_
|
|
 |
98.7 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:d-lysine 5,6-aminomutase beta subunit;
PDBTitle: crystal structure of lysine 5,6-aminomutase in complex with plp,2 cobalamin, and 5'-deoxyadenosine
|
|
|
|
| 14 | d1fmfa_
|
|
 |
98.6 |
10 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
|
| 15 | c1y80A_
|
|
 |
98.6 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted cobalamin binding protein;
PDBTitle: structure of a corrinoid (factor iiim)-binding protein from moorella2 thermoacetica
|
|
|
|
| 16 | c4r3uD_
|
|
 |
98.6 |
19 |
PDB header:isomerase
Chain: D: PDB Molecule:2-hydroxyisobutyryl-coa mutase small subunit;
PDBTitle: crystal structure of 2-hydroxyisobutyryl-coa mutase
|
|
|
|
| 17 | c4jgiB_
|
|
 |
98.6 |
19 |
PDB header:protein binding
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: 1.5 angstrom crystal structure of a novel cobalamin-binding protein2 from desulfitobacterium hafniense dcb-2
|
|
|
|
| 18 | c1bmtB_
|
|
 |
98.5 |
13 |
PDB header:methyltransferase
Chain: B: PDB Molecule:methionine synthase;
PDBTitle: how a protein binds b12: a 3.o angstrom x-ray structure of2 the b12-binding domains of methionine synthase
|
|
|
|
| 19 | d3bula2
|
|
 |
98.5 |
13 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
|
| 20 | c2i2xD_
|
|
 |
98.4 |
10 |
PDB header:transferase
Chain: D: PDB Molecule:methyltransferase 1;
PDBTitle: crystal structure of methanol:cobalamin methyltransferase complex2 mtabc from methanosarcina barkeri
|
|
|
|
| 21 | c3ezxA_ |
|
not modelled |
98.3 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:monomethylamine corrinoid protein 1;
PDBTitle: structure of methanosarcina barkeri monomethylamine2 corrinoid protein
|
|
|
| 22 | c1k98A_ |
|
not modelled |
98.1 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:methionine synthase;
PDBTitle: adomet complex of meth c-terminal fragment
|
|
|
| 23 | c4hh3C_ |
|
not modelled |
97.9 |
15 |
PDB header:flavoprotein/transcription
Chain: C: PDB Molecule:appa protein;
PDBTitle: structure of the appa-ppsr2 core complex from rb. sphaeroides
|
|
|
| 24 | d1ccwb_ |
|
not modelled |
97.8 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Cobalamin (vitamin B12)-dependent enzymes
Family:Glutamate mutase, large subunit |
|
|
| 25 | c3whpA_ |
|
not modelled |
97.1 |
20 |
PDB header:gene regulation
Chain: A: PDB Molecule:probable transcriptional regulator;
PDBTitle: crystal structure of the c-terminal domain of themus thermophilus litr2 in complex with cobalamin
|
|
|
| 26 | c5c8eC_ |
|
not modelled |
96.8 |
20 |
PDB header:transcription regulator/dna
Chain: C: PDB Molecule:light-dependent transcriptional regulator carh;
PDBTitle: crystal structure of thermus thermophilus carh bound to2 adenosylcobalamin and a 26-bp dna segment
|
|
|
| 27 | c3nhzA_ |
|
not modelled |
94.8 |
17 |
PDB header:dna binding protein
Chain: A: PDB Molecule:two component system transcriptional regulator mtra;
PDBTitle: structure of n-terminal domain of mtra
|
|
|
| 28 | d1zesa1 |
|
not modelled |
94.6 |
7 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 29 | d1ys7a2 |
|
not modelled |
93.8 |
19 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 30 | c2mswA_ |
|
not modelled |
92.6 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:response regulator/sensor histidine kinase;
PDBTitle: ligand-induced folding of a receiver domain
|
|
|
| 31 | c4hh0B_ |
|
not modelled |
92.5 |
15 |
PDB header:flavoprotein,signaling protein
Chain: B: PDB Molecule:appa protein;
PDBTitle: dark-state structure of appa c20s without the cys-rich region from rb.2 sphaeroides
|
|
|
| 32 | c1ys7B_ |
|
not modelled |
91.2 |
20 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulatory protein prra;
PDBTitle: crystal structure of the response regulator protein prra complexed2 with mg2+
|
|
|
| 33 | c3ktoA_ |
|
not modelled |
91.1 |
9 |
PDB header:transcription regulator
Chain: A: PDB Molecule:response regulator receiver protein;
PDBTitle: crystal structure of response regulator receiver protein2 from pseudoalteromonas atlantica
|
|
|
| 34 | d1yioa2 |
|
not modelled |
91.0 |
18 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 35 | d1mvoa_ |
|
not modelled |
90.5 |
13 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 36 | c5iejA_ |
|
not modelled |
89.5 |
18 |
PDB header:protein
Chain: A: PDB Molecule:sdrg;
PDBTitle: solution structure of the bef3-activated conformation of sdrg from2 pseudomonas melonis fr1
|
|
|
| 37 | c3a0rB_ |
|
not modelled |
89.3 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:response regulator;
PDBTitle: crystal structure of histidine kinase thka (tm1359) in complex with2 response regulator protein trra (tm1360)
|
|
|
| 38 | d1krwa_ |
|
not modelled |
88.4 |
12 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 39 | c5tqjA_ |
|
not modelled |
88.0 |
16 |
PDB header:signaling protein
Chain: A: PDB Molecule:response regulator receiver protein;
PDBTitle: crystal structure of response regulator receiver protein from2 burkholderia phymatum
|
|
|
| 40 | c2qzjC_ |
|
not modelled |
86.4 |
12 |
PDB header:transcription
Chain: C: PDB Molecule:two-component response regulator;
PDBTitle: crystal structure of a two-component response regulator from2 clostridium difficile
|
|
|
| 41 | c6ifhA_ |
|
not modelled |
85.8 |
8 |
PDB header:transferase
Chain: A: PDB Molecule:sporulation initiation phosphotransferase f;
PDBTitle: unphosphorylated spo0f from paenisporosarcina sp. tg-14
|
|
|
| 42 | c1zn2A_ |
|
not modelled |
85.3 |
19 |
PDB header:transcription regulator
Chain: A: PDB Molecule:response regulatory protein;
PDBTitle: low resolution structure of response regulator styr
|
|
|
| 43 | d1xhfa1 |
|
not modelled |
84.6 |
10 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 44 | c5e3jB_ |
|
not modelled |
84.2 |
12 |
PDB header:transcription
Chain: B: PDB Molecule:response regulator rsta;
PDBTitle: the response regulator rsta is a potential drug target for2 acinetobacter baumannii
|
|
|
| 45 | c2zayA_ |
|
not modelled |
83.8 |
14 |
PDB header:signaling protein
Chain: A: PDB Molecule:response regulator receiver protein;
PDBTitle: crystal structure of response regulator from desulfuromonas2 acetoxidans
|
|
|
| 46 | c4qpjC_ |
|
not modelled |
83.8 |
11 |
PDB header:signaling protein/dna binding protein
Chain: C: PDB Molecule:cell cycle response regulator ctra;
PDBTitle: 2.7 angstrom structure of a phosphotransferase in complex with a2 receiver domain
|
|
|
| 47 | c5yaaD_ |
|
not modelled |
83.3 |
15 |
PDB header:hydrolase
Chain: D: PDB Molecule:meiosis regulator and mrna stability factor 1;
PDBTitle: crystal structure of marf1 nyn domain from mus musculus
|
|
|
| 48 | d1wu7a1 |
|
not modelled |
81.9 |
16 |
Fold:Anticodon-binding domain-like
Superfamily:Class II aaRS ABD-related
Family:Anticodon-binding domain of Class II aaRS |
|
|
| 49 | c5ulbA_ |
|
not modelled |
81.8 |
14 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:putative sugar abc transporter;
PDBTitle: crystal structure of sugar abc transporter from yersinia2 enterocolitica subsp. enterocolitica 8081
|
|
|
| 50 | d2a9pa1 |
|
not modelled |
81.5 |
10 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 51 | d1zh2a1 |
|
not modelled |
81.1 |
14 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 52 | c5ajtA_ |
|
not modelled |
80.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphoribohydrolase lonely guy;
PDBTitle: crystal structure of ligand-free phosphoribohydrolase lonely guy from2 claviceps purpurea
|
|
|
| 53 | c2h51B_ |
|
not modelled |
80.0 |
29 |
PDB header:hydrolase/hydrolase inhibitor
Chain: B: PDB Molecule:caspase-1;
PDBTitle: crystal structure of human caspase-1 (glu390->asp and arg286->lys) in2 complex with 3-[2-(2-benzyloxycarbonylamino-3-methyl-butyrylamino)-3 propionylamino]-4-oxo-pentanoic acid (z-vad-fmk)
|
|
|
| 54 | c4b09F_ |
|
not modelled |
79.6 |
9 |
PDB header:transcription
Chain: F: PDB Molecule:transcriptional regulatory protein baer;
PDBTitle: structure of unphosphorylated baer dimer
|
|
|
| 55 | c3r0jA_ |
|
not modelled |
79.4 |
18 |
PDB header:dna binding protein
Chain: A: PDB Molecule:possible two component system response transcriptional
PDBTitle: structure of phop from mycobacterium tuberculosis
|
|
|
| 56 | c2c2zB_ |
|
not modelled |
79.1 |
33 |
PDB header:hydrolase/hydrolase inhibitor
Chain: B: PDB Molecule:caspase-8 p10 subunit;
PDBTitle: crystal structure of caspase-8 in complex with aza-peptide michael2 acceptor inhibitor
|
|
|
| 57 | c5x5jA_ |
|
not modelled |
79.1 |
12 |
PDB header:dna binding protein
Chain: A: PDB Molecule:ader;
PDBTitle: crystal structure of response regulator ader receiver domain
|
|
|
| 58 | c3w9sB_ |
|
not modelled |
78.8 |
11 |
PDB header:signaling protein/antimicrobial protein
Chain: B: PDB Molecule:ompr family response regulator in two-component regulatory
PDBTitle: crystal structure analysis of the n-terminal receiver domain of2 response regulator pmra
|
|
|
| 59 | c2c1eB_ |
|
not modelled |
78.7 |
33 |
PDB header:hydrolase/hydrolase inhibitor
Chain: B: PDB Molecule:caspase-3 subunit p12;
PDBTitle: crystal structures of caspase-3 in complex with aza-peptide michael2 acceptor inhibitors.
|
|
|
| 60 | c3cfyA_ |
|
not modelled |
78.6 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative luxo repressor protein;
PDBTitle: crystal structure of signal receiver domain of putative luxo repressor2 protein from vibrio parahaemolyticus
|
|
|
| 61 | c2jk1A_ |
|
not modelled |
78.6 |
17 |
PDB header:dna-binding
Chain: A: PDB Molecule:hydrogenase transcriptional regulatory protein hupr1;
PDBTitle: crystal structure of the wild-type hupr receiver domain
|
|
|
| 62 | d1peya_ |
|
not modelled |
78.4 |
5 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 63 | c2zwmA_ |
|
not modelled |
78.4 |
11 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulatory protein yycf;
PDBTitle: crystal structure of yycf receiver domain from bacillus2 subtilis
|
|
|
| 64 | c2qljB_ |
|
not modelled |
76.6 |
38 |
PDB header:hydrolase/hydrolase inhibitor
Chain: B: PDB Molecule:caspase-7;
PDBTitle: crystal structure of caspase-7 with inhibitor ac-wehd-cho
|
|
|
| 65 | c3c3mA_ |
|
not modelled |
76.5 |
13 |
PDB header:signaling protein
Chain: A: PDB Molecule:response regulator receiver protein;
PDBTitle: crystal structure of the n-terminal domain of response regulator2 receiver protein from methanoculleus marisnigri jr1
|
|
|
| 66 | c3hdgE_ |
|
not modelled |
76.0 |
11 |
PDB header:structural genomics, unknown function
Chain: E: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the n-terminal domain of an2 uncharacterized protein (ws1339) from wolinella3 succinogenes
|
|
|
| 67 | c3jteA_ |
|
not modelled |
75.6 |
15 |
PDB header:protein binding
Chain: A: PDB Molecule:response regulator receiver protein;
PDBTitle: crystal structure of response regulator receiver domain protein from2 clostridium thermocellum
|
|
|
| 68 | c3snkA_ |
|
not modelled |
75.1 |
14 |
PDB header:signaling protein
Chain: A: PDB Molecule:response regulator chey-like protein;
PDBTitle: crystal structure of a response regulator chey-like protein (mll6475)2 from mesorhizobium loti at 2.02 a resolution
|
|
|
| 69 | c5uicA_ |
|
not modelled |
74.8 |
12 |
PDB header:transcription
Chain: A: PDB Molecule:two-component response regulator;
PDBTitle: structure of the francisella response regulator receiver domain, qseb
|
|
|
| 70 | c3f6cB_ |
|
not modelled |
74.2 |
13 |
PDB header:dna binding protein
Chain: B: PDB Molecule:positive transcription regulator evga;
PDBTitle: crystal structure of n-terminal domain of positive transcription2 regulator evga from escherichia coli
|
|
|
| 71 | c2hqrA_ |
|
not modelled |
73.3 |
11 |
PDB header:signaling protein
Chain: A: PDB Molecule:putative transcriptional regulator;
PDBTitle: structure of a atypical orphan response regulator protein revealed a2 new phosphorylation-independent regulatory mechanism
|
|
|
| 72 | d1ny5a1 |
|
not modelled |
73.0 |
10 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 73 | c4ldaF_ |
|
not modelled |
71.9 |
20 |
PDB header:transcription
Chain: F: PDB Molecule:tadz;
PDBTitle: crystal structure of a chey-like protein (tadz) from pseudomonas2 aeruginosa pao1 at 2.70 a resolution
|
|
|
| 74 | c2qr3A_ |
|
not modelled |
71.6 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:two-component system response regulator;
PDBTitle: crystal structure of the n-terminal signal receiver domain of two-2 component system response regulator from bacteroides fragilis
|
|
|
| 75 | c5zbjA_ |
|
not modelled |
71.2 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative cytokinin riboside 5'-monophosphate
PDBTitle: crystal strcuture of type-i log from pseudomonas aeruginosa pao1
|
|
|
| 76 | c3sipB_ |
|
not modelled |
71.0 |
33 |
PDB header:hydrolase/ligase/hydrolase
Chain: B: PDB Molecule:caspase;
PDBTitle: crystal structure of drice and diap1-bir1 complex
|
|
|
| 77 | d1u0sy_ |
|
not modelled |
70.6 |
10 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 78 | c2qxyB_ |
|
not modelled |
70.6 |
11 |
PDB header:transcription
Chain: B: PDB Molecule:response regulator;
PDBTitle: crystal structure of a response regulator from thermotoga2 maritima
|
|
|
| 79 | c3p45F_ |
|
not modelled |
70.3 |
29 |
PDB header:hydrolase
Chain: F: PDB Molecule:caspase-6;
PDBTitle: crystal structure of apo-caspase-6 at physiological ph
|
|
|
| 80 | d3clsc1 |
|
not modelled |
69.9 |
16 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ETFP subunits |
|
|
| 81 | c2q4oA_ |
|
not modelled |
69.7 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein at2g37210/t2n18.3;
PDBTitle: ensemble refinement of the crystal structure of a lysine2 decarboxylase-like protein from arabidopsis thaliana gene at2g37210
|
|
|
| 82 | d2q4oa1 |
|
not modelled |
69.7 |
22 |
Fold:MCP/YpsA-like
Superfamily:MCP/YpsA-like
Family:MoCo carrier protein-like |
|
|
| 83 | c2p2cD_ |
|
not modelled |
69.3 |
33 |
PDB header:hydrolase
Chain: D: PDB Molecule:caspase-2;
PDBTitle: inhibition of caspase-2 by a designed ankyrin repeat protein (darpin)
|
|
|
| 84 | c3hdvB_ |
|
not modelled |
69.2 |
18 |
PDB header:transcription regulator
Chain: B: PDB Molecule:response regulator;
PDBTitle: crystal structure of response regulator receiver protein from2 pseudomonas putida
|
|
|
| 85 | c5ow0B_ |
|
not modelled |
68.5 |
9 |
PDB header:electron transport
Chain: B: PDB Molecule:electron transfer flavoprotein, beta subunit;
PDBTitle: crystal structure of an electron transfer flavoprotein from2 geobacillus metallireducens
|
|
|
| 86 | c2nt3A_ |
|
not modelled |
67.8 |
14 |
PDB header:signaling protein
Chain: A: PDB Molecule:response regulator homolog;
PDBTitle: receiver domain from myxococcus xanthus social motility protein frzs2 (y102a mutant)
|
|
|
| 87 | c2lpmA_ |
|
not modelled |
67.1 |
17 |
PDB header:transcription regulator
Chain: A: PDB Molecule:two-component response regulator;
PDBTitle: chemical shift and structure assignments for sma0114
|
|
|
| 88 | c4rweA_ |
|
not modelled |
67.0 |
17 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:sugar-binding transport protein;
PDBTitle: the crystal structure of a sugar-binding transport protein from2 yersinia pestis co92
|
|
|
| 89 | d1ydha_ |
|
not modelled |
66.6 |
20 |
Fold:MCP/YpsA-like
Superfamily:MCP/YpsA-like
Family:MoCo carrier protein-like |
|
|
| 90 | d1x94a_ |
|
not modelled |
66.1 |
14 |
Fold:SIS domain
Superfamily:SIS domain
Family:mono-SIS domain |
|
|
| 91 | c2j48A_ |
|
not modelled |
65.8 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:two-component sensor kinase;
PDBTitle: nmr structure of the pseudo-receiver domain of the cika protein.
|
|
|
| 92 | d1t35a_ |
|
not modelled |
65.7 |
19 |
Fold:MCP/YpsA-like
Superfamily:MCP/YpsA-like
Family:MoCo carrier protein-like |
|
|
| 93 | c4h60A_ |
|
not modelled |
65.6 |
18 |
PDB header:signaling protein
Chain: A: PDB Molecule:chemotaxis protein chey;
PDBTitle: high resolution structure of vibrio cholerae chemotaxis protein chey42 crystallized in low ph (4.0) condition
|
|
|
| 94 | c3trjC_ |
|
not modelled |
65.1 |
15 |
PDB header:isomerase
Chain: C: PDB Molecule:phosphoheptose isomerase;
PDBTitle: structure of a phosphoheptose isomerase from francisella tularensis
|
|
|
| 95 | c2qipA_ |
|
not modelled |
65.1 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein of unknown function vpa0982;
PDBTitle: crystal structure of a protein of unknown function vpa0982 from vibrio2 parahaemolyticus rimd 2210633
|
|
|
| 96 | c3hv2B_ |
|
not modelled |
64.2 |
15 |
PDB header:signaling protein
Chain: B: PDB Molecule:response regulator/hd domain protein;
PDBTitle: crystal structure of signal receiver domain of hd domain-containing2 protein from pseudomonas fluorescens pf-5
|
|
|
| 97 | d1qkka_ |
|
not modelled |
64.1 |
12 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 98 | c3rqiA_ |
|
not modelled |
64.0 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:response regulator protein;
PDBTitle: crystal structure of a response regulator protein from burkholderia2 pseudomallei with a phosphorylated aspartic acid, calcium ion and3 citrate
|
|
|
| 99 | c3sbxC_ |
|
not modelled |
63.2 |
26 |
PDB header:unknown function
Chain: C: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of a putative uncharacterized protein from2 mycobacterium marinum bound to adenosine 5'-monophosphate amp
|
|
|
| 100 | d1cmwa2 |
|
not modelled |
63.2 |
14 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
|
|
| 101 | d2d69a1 |
|
not modelled |
62.5 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 102 | c3grcD_ |
|
not modelled |
62.5 |
13 |
PDB header:transferase
Chain: D: PDB Molecule:sensor protein, kinase;
PDBTitle: crystal structure of a sensor protein from polaromonas sp.2 js666
|
|
|
| 103 | d1p6qa_ |
|
not modelled |
62.4 |
11 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 104 | c5itsD_ |
|
not modelled |
62.2 |
26 |
PDB header:hydrolase
Chain: D: PDB Molecule:cytokinin riboside 5'-monophosphate phosphoribohydrolase;
PDBTitle: crystal strcuture of log from corynebacterium glutamicum
|
|
|
| 105 | c4ggmX_ |
|
not modelled |
62.1 |
24 |
PDB header:hydrolase
Chain: X: PDB Molecule:udp-2,3-diacylglucosamine pyrophosphatase lpxi;
PDBTitle: structure of lpxi
|
|
|
| 106 | c2rjnA_ |
|
not modelled |
61.8 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:response regulator receiver:metal-dependent
PDBTitle: crystal structure of an uncharacterized protein q2bku2 from2 neptuniibacter caesariensis
|
|
|
| 107 | c3gt7A_ |
|
not modelled |
61.6 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:sensor protein;
PDBTitle: crystal structure of signal receiver domain of signal transduction2 histidine kinase from syntrophus aciditrophicus
|
|
|
| 108 | c3sirD_ |
|
not modelled |
60.9 |
33 |
PDB header:hydrolase
Chain: D: PDB Molecule:caspase;
PDBTitle: crystal structure of drice
|
|
|
| 109 | d2ayxa1 |
|
not modelled |
60.8 |
13 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 110 | c5e5nD_ |
|
not modelled |
60.6 |
13 |
PDB header:hydrolase
Chain: D: PDB Molecule:polyketide synthase pksl;
PDBTitle: ketosynthase from module 6 of the bacillaene synthase from bacillus2 subtilis 168 (c167s mutant, crystal form 1)
|
|
|
| 111 | c2fp3A_ |
|
not modelled |
60.4 |
20 |
PDB header:hydrolysis/apoptosis
Chain: A: PDB Molecule:caspase nc;
PDBTitle: crystal structure of the drosophila initiator caspase dronc
|
|
|
| 112 | c4q7eA_ |
|
not modelled |
60.4 |
9 |
PDB header:signaling protein
Chain: A: PDB Molecule:response regulator of a two component regulatory system;
PDBTitle: non-phosphorylated hemr receiver domain from leptospira biflexa
|
|
|
| 113 | c4d6yA_ |
|
not modelled |
60.2 |
11 |
PDB header:signaling protein
Chain: A: PDB Molecule:bacterial regulatory, fis family protein;
PDBTitle: crystal structure of the receiver domain of ntrx from2 brucella abortus in complex with beryllofluoride and3 magnesium
|
|
|
| 114 | c5t3yA_ |
|
not modelled |
59.9 |
15 |
PDB header:signaling protein
Chain: A: PDB Molecule:two-component system response regulator;
PDBTitle: solution structure of response regulator protein from burkholderia2 multivorans
|
|
|
| 115 | d1jx6a_ |
|
not modelled |
58.4 |
14 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
|
|
| 116 | c3lteH_ |
|
not modelled |
57.8 |
12 |
PDB header:transcription
Chain: H: PDB Molecule:response regulator;
PDBTitle: crystal structure of response regulator (signal receiver domain) from2 bermanella marisrubri
|
|
|
| 117 | c3quaA_ |
|
not modelled |
57.5 |
26 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of a putative uncharacterized protein and possible2 molybdenum cofactor protein from mycobacterium smegmatis
|
|
|
| 118 | d1jbea_ |
|
not modelled |
56.8 |
15 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 119 | c1ny5A_ |
|
not modelled |
56.3 |
10 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator (ntrc family);
PDBTitle: crystal structure of sigm54 activator (aaa+ atpase) in the inactive2 state
|
|
|
| 120 | d1qe0a1 |
|
not modelled |
55.5 |
15 |
Fold:Anticodon-binding domain-like
Superfamily:Class II aaRS ABD-related
Family:Anticodon-binding domain of Class II aaRS |
|
|