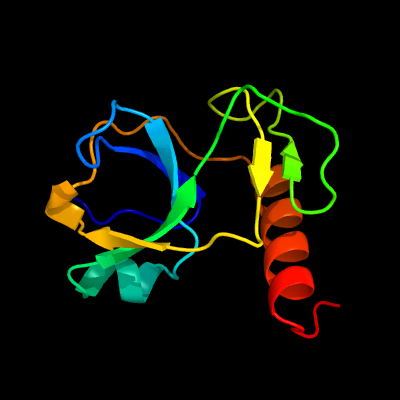
1 c5xe3B_
100.0
97
PDB header: hydrolase/antitoxinChain: B: PDB Molecule: endoribonuclease mazf4;PDBTitle: endoribonuclease in complex with its cognate antitoxin from2 mycobacterial species
2 d1ub4a_
100.0
22
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: Kid/PemK
3 c5wygC_
100.0
26
PDB header: hydrolaseChain: C: PDB Molecule: probable endoribonuclease mazf7;PDBTitle: the crystal structure of the apo form of mtb mazf
4 d1m1fa_
100.0
33
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: Kid/PemK
5 d1ne8a_
100.0
24
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: Kid/PemK
6 c4mzpC_
100.0
29
PDB header: hydrolaseChain: C: PDB Molecule: mazf mrna interferase;PDBTitle: mazf from s. aureus crystal form iii, c2221, 2.7 a
7 c5hk3B_
99.9
31
PDB header: hydrolase/dnaChain: B: PDB Molecule: endoribonuclease mazf6;PDBTitle: crystal structure of m. tuberculosis mazf-mt3 t52d-f62d mutant in2 complex with dna
8 c5hjzA_
99.9
21
PDB header: hydrolase/rnaChain: A: PDB Molecule: endoribonuclease mazf9;PDBTitle: structure of m. tuberculosis mazf-mt1 (rv2801c) in complex with rna
9 c5ccaA_
99.9
26
PDB header: hydrolaseChain: A: PDB Molecule: endoribonuclease mazf3;PDBTitle: crystal structure of mtb toxin
10 d3vuba_
95.9
11
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: CcdB
11 c3jrzA_
95.9
11
PDB header: toxinChain: A: PDB Molecule: ccdb;PDBTitle: ccdbvfi-formii-ph5.6
12 c5ikjA_
57.9
28
PDB header: transcriptionChain: A: PDB Molecule: cryptic loci regulator 2;PDBTitle: structure of clr2 bound to the clr1 c-terminus
13 c3llrA_
25.2
22
PDB header: transferaseChain: A: PDB Molecule: dna (cytosine-5)-methyltransferase 3a;PDBTitle: crystal structure of the pwwp domain of human dna (cytosine-5-)-2 methyltransferase 3 alpha
14 d1h3za_
21.6
12
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: PWWP domain
15 c2l89A_
19.4
16
PDB header: protein bindingChain: A: PDB Molecule: pwwp domain-containing protein 1;PDBTitle: solution structure of pdp1 pwwp domain reveals its unique binding2 sites for methylated h4k20 and dna
16 c4me8A_
18.3
9
PDB header: hydrolaseChain: A: PDB Molecule: signal peptidase i;PDBTitle: crystal structure of a signal peptidase i (ef3073) from enterococcus2 faecalis v583 at 2.27 a resolution
17 c2z3tD_
18.0
19
PDB header: oxidoreductaseChain: D: PDB Molecule: cytochrome p450;PDBTitle: crystal structure of substrate free cytochrome p450 stap2 (cyp245a1)
18 c4rmoA_
9.8
12
PDB header: toxin/rnaChain: A: PDB Molecule: cptn toxin;PDBTitle: crystal structure of the cptin type iii toxin-antitoxin system from2 eubacterium rectale
19 c4z5qA_
9.2
29
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome p450 hydroxylase;PDBTitle: crystal structure of the lnmz cytochrome p450 hydroxylase from the2 leinamycin biosynthetic pathway of streptomyces atroolivaceus s-1403 at 1.8 a resolution
20 c2dkkA_
9.2
18
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome p450;PDBTitle: structure/function studies of cytochrome p450 158a1 from streptomyces2 coelicolor a3(2)
21 d1rz4a1
not modelled
7.9
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Eukaryotic translation initiation factor 3 subunit 12, eIF3k, C-terminal domain
22 c1pjtB_
not modelled
7.9
31
PDB header: transferase/oxidoreductase/lyaseChain: B: PDB Molecule: siroheme synthase;PDBTitle: the structure of the ser128ala point-mutant variant of cysg, the2 multifunctional methyltransferase/dehydrogenase/ferrochelatase for3 siroheme synthesis
23 d1okja2
not modelled
7.4
30
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: YeaZ-like
24 c2of3A_
not modelled
6.6
8
PDB header: structural protein, cell cycleChain: A: PDB Molecule: zyg-9;PDBTitle: tog domain structure from c.elegans zyg9
25 c3zeyM_
not modelled
6.6
22
PDB header: ribosomeChain: M: PDB Molecule: 40s ribosomal protein s18, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
26 c4xe3B_
not modelled
6.6
27
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome p-450;PDBTitle: olep, the cytochrome p450 epoxidase from streptomyces antibioticus2 involved in oleandomycin biosynthesis: functional analysis and3 crystallographic structure in complex with clotrimazole.
27 d1s1fa_
not modelled
6.5
27
Fold: Cytochrome P450Superfamily: Cytochrome P450Family: Cytochrome P450
28 c1bdsA_
not modelled
6.1
43
PDB header: anti-hypertensive, anti-viral proteinChain: A: PDB Molecule: bds-i;PDBTitle: determination of the three-dimensional solution structure of the2 antihypertensive and antiviral protein bds-i from the sea anemone3 anemonia sulcata. a study using nuclear magnetic resonance and hybrid4 distance geometry-dynamical simulated annealing
29 d1bdsa_
not modelled
6.1
43
Fold: Defensin-likeSuperfamily: Defensin-likeFamily: Defensin
30 c2zkqm_
not modelled
6.1
19
PDB header: ribosomal protein/rnaChain: M: PDB Molecule: PDBTitle: structure of a mammalian ribosomal 40s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
31 c5zc2B_
not modelled
6.0
8
PDB header: flavoproteinChain: B: PDB Molecule: p-hydroxyphenylacetate 3-hydroxylase, reductase component;PDBTitle: acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (hpah),2 reductase component (c1)
32 c1qp6B_
not modelled
6.0
43
PDB header: de novo proteinChain: B: PDB Molecule: protein (alpha2d);PDBTitle: solution structure of alpha2d
33 d2daqa1
not modelled
5.8
9
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: PWWP domain
34 c1cffB_
not modelled
5.4
43
PDB header: calmodulinChain: B: PDB Molecule: calcium pump;PDBTitle: nmr solution structure of a complex of calmodulin with a binding2 peptide of the ca2+-pump
35 c3iz6M_
not modelled
5.4
15
PDB header: ribosomeChain: M: PDB Molecule: 40s ribosomal protein s18 (s13p);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
36 c2gfuA_
not modelled
5.3
17
PDB header: dna binding proteinChain: A: PDB Molecule: dna mismatch repair protein msh6;PDBTitle: nmr solution structure of the pwwp domain of mismatch2 repair protein hmsh6