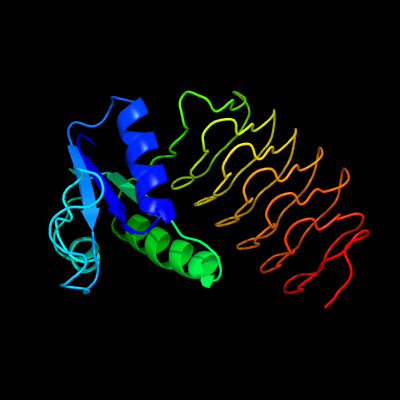
| 1 | c4m98A_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:pilin glycosylation protein;
PDBTitle: acetyltransferase domain of pglb from neisseria gonorrhoeae fa1090
|
|
|
|
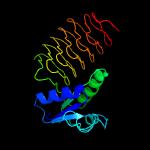
| 2 | c4m9cC_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: C: PDB Molecule:bacterial transferase hexapeptide (three repeats) family
PDBTitle: weei from acinetobacter baumannii aye
|
|
|
|
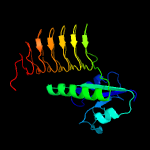
| 3 | c4ea8A_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:perosamine n-acetyltransferase;
PDBTitle: x-ray crystal structure of perb from caulobacter crescentus in complex2 with coenzyme a and gdp-n-acetylperosamine at 1 angstrom resolution
|
|
|
|
| 4 | d3bswa1
|
|
 |
100.0 |
20 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:PglD-like |
|
|
|
| 5 | c2iu9C_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: C: PDB Molecule:udp-3-o-[3-hydroxymyristoyl] glucosamine
PDBTitle: chlamydia trachomatis lpxd with 100mm udpglcnac (complex ii)
|
|
|
|
| 6 | c3cj8B_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:2,3,4,5-tetrahydropyridine-2,6-dicarboxylate n-
PDBTitle: crystal structure of 2,3,4,5-tetrahydropyridine-2-carboxylate n-2 succinyltransferase from enterococcus faecalis v583
|
|
|
|
| 7 | c4e6tA_
|
|
 |
100.0 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:acyl-[acyl-carrier-protein]--udp-n-acetylglucosamine o-
PDBTitle: structure of lpxa from acinetobacter baumannii at 1.8a resolution2 (p212121 form)
|
|
|
|
| 8 | c3eh0C_
|
|
 |
100.0 |
25 |
PDB header:transferase
Chain: C: PDB Molecule:udp-3-o-[3-hydroxymyristoyl] glucosamine n-
PDBTitle: crystal structure of lpxd from escherichia coli
|
|
|
|
| 9 | c4eqyC_
|
|
 |
100.0 |
27 |
PDB header:transferase
Chain: C: PDB Molecule:acyl-[acyl-carrier-protein]--udp-n-acetylglucosamine o-
PDBTitle: crystal structure of acyl-[acyl-carrier-protein]--udp-n-2 acetylglucosamine o-acyltransferase from burkholderia thailandensis
|
|
|
|
| 10 | c3i3aC_
|
|
 |
100.0 |
23 |
PDB header:transferase
Chain: C: PDB Molecule:acyl-[acyl-carrier-protein]--udp-n-
PDBTitle: structural basis for the sugar nucleotide and acyl chain2 selectivity of leptospira interrogans lpxa
|
|
|
|
| 11 | c4r36A_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:putative acyl-[acyl-carrier-protein]--udp-n-
PDBTitle: crystal structure analysis of lpxa, a udp-n-acetylglucosamine2 acyltransferase from bacteroides fragilis 9343
|
|
|
|
| 12 | c3r0sA_
|
|
 |
100.0 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:acyl-[acyl-carrier-protein]--udp-n-acetylglucosamine o-
PDBTitle: udp-n-acetylglucosamine acyltransferase from campylobacter jejuni
|
|
|
|
| 13 | c3pmoA_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:udp-3-o-[3-hydroxymyristoyl] glucosamine n-acyltransferase;
PDBTitle: the structure of lpxd from pseudomonas aeruginosa at 1.3 a resolution
|
|
|
|
| 14 | c5jxxC_
|
|
 |
100.0 |
23 |
PDB header:transferase
Chain: C: PDB Molecule:acyl-[acyl-carrier-protein]--udp-n-acetylglucosamine o-
PDBTitle: crystal structure of udp-n-acetylglucosamine o-acyltransferase (lpxa)2 from moraxella catarrhalis rh4.
|
|
|
|
| 15 | c5f42B_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:acyl-[acyl-carrier-protein]--udp-n-acetylglucosamine o-
PDBTitle: activity and crystal structure of francisella novicida udp-n-2 acetylglucosamine acyltransferase
|
|
|
|
| 16 | d2jf2a1
|
|
 |
100.0 |
25 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:UDP N-acetylglucosamine acyltransferase |
|
|
|
| 17 | d1j2za_
|
|
 |
100.0 |
21 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:UDP N-acetylglucosamine acyltransferase |
|
|
|
| 18 | c3t57A_
|
|
 |
100.0 |
32 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine o-acyltransferase domain-containing
PDBTitle: activity and crystal structure of arabidopsis udp-n-acetylglucosamine2 acyltransferase
|
|
|
|
| 19 | c4e75A_
|
|
 |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:udp-3-o-acylglucosamine n-acyltransferase;
PDBTitle: structure of lpxd from acinetobacter baumannii at 2.85a resolution2 (p21 form)
|
|
|
|
| 20 | c5dg3D_
|
|
 |
100.0 |
25 |
PDB header:transferase
Chain: D: PDB Molecule:acyl-[acyl-carrier-protein]--udp-n-acetylglucosamine o-
PDBTitle: structure of pseudomonas aeruginosa lpxa in complex with udp-3-o-(r-3-2 hydroxydecanoyl)-glcnac
|
|
|
|
| 21 | c3fttA_ |
|
not modelled |
100.0 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:putative acetyltransferase sacol2570;
PDBTitle: crystal structure of the galactoside o-acetyltransferase from2 staphylococcus aureus
|
|
|
| 22 | c3srtB_ |
|
not modelled |
100.0 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:maltose o-acetyltransferase;
PDBTitle: the crystal structure of a maltose o-acetyltransferase from2 clostridium difficile 630
|
|
|
| 23 | d1krra_ |
|
not modelled |
100.0 |
19 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Galactoside acetyltransferase-like |
|
|
| 24 | c4mzuG_ |
|
not modelled |
100.0 |
26 |
PDB header:isomerase, transferase
Chain: G: PDB Molecule:wxcm-like protein;
PDBTitle: crystal structure of fdtd, a bifunctional ketoisomerase/n-2 acetyltransferase from shewanella denitrificans
|
|
|
| 25 | c3ectA_ |
|
not modelled |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:hexapeptide-repeat containing-acetyltransferase;
PDBTitle: crystal structure of the hexapeptide-repeat containing-2 acetyltransferase vca0836 from vibrio cholerae
|
|
|
| 26 | c2ic7A_ |
|
not modelled |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:maltose transacetylase;
PDBTitle: crystal structure of maltose transacetylase from geobacillus2 kaustophilus
|
|
|
| 27 | c3r8yD_ |
|
not modelled |
100.0 |
23 |
PDB header:transferase
Chain: D: PDB Molecule:2,3,4,5-tetrahydropyridine-2,6-dicarboxylate n-
PDBTitle: structure of the bacillus anthracis tetrahydropicolinate2 succinyltransferase
|
|
|
| 28 | d1ocxa_ |
|
not modelled |
99.9 |
20 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Galactoside acetyltransferase-like |
|
|
| 29 | c3fsbB_ |
|
not modelled |
99.9 |
29 |
PDB header:transferase
Chain: B: PDB Molecule:qdtc;
PDBTitle: crystal structure of qdtc, the dtdp-3-amino-3,6-dideoxy-d-2 glucose n-acetyl transferase from thermoanaerobacterium3 thermosaccharolyticum in complex with coa and dtdp-3-amino-4 quinovose
|
|
|
| 30 | c3mqhD_ |
|
not modelled |
99.9 |
23 |
PDB header:transferase
Chain: D: PDB Molecule:lipopolysaccharides biosynthesis acetyltransferase;
PDBTitle: crystal structure of the 3-n-acetyl transferase wlbb from bordetella2 petrii in complex with coa and udp-3-amino-2-acetamido-2,3-dideoxy3 glucuronic acid
|
|
|
| 31 | c3vbnA_ |
|
not modelled |
99.9 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:galactoside o-acetyltransferase;
PDBTitle: crystal structure of the d94a mutant of antd, an n-acyltransferase2 from bacillus cereus in complex with dtdp and coenzyme a
|
|
|
| 32 | d1t3da_ |
|
not modelled |
99.9 |
16 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Serine acetyltransferase |
|
|
| 33 | c1t3dB_ |
|
not modelled |
99.9 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:serine acetyltransferase;
PDBTitle: crystal structure of serine acetyltransferase from e.coli at 2.2a
|
|
|
| 34 | d1mr7a_ |
|
not modelled |
99.9 |
15 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Galactoside acetyltransferase-like |
|
|
| 35 | c4e8lC_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: C: PDB Molecule:virginiamycin a acetyltransferase;
PDBTitle: crystal structure of streptogramin group a antibiotic2 acetyltransferase vata from staphylococcus aureus
|
|
|
| 36 | c6mfkA_ |
|
not modelled |
99.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:chloramphenicol acetyltransferase;
PDBTitle: crystal structure of chloramphenicol acetyltransferase from2 elizabethkingia anophelis
|
|
|
| 37 | d1g97a1 |
|
not modelled |
99.9 |
25 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:GlmU C-terminal domain-like |
|
|
| 38 | d1ssqa_ |
|
not modelled |
99.9 |
13 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Serine acetyltransferase |
|
|
| 39 | c3jqyB_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:polysialic acid o-acetyltransferase;
PDBTitle: crystal strucutre of the polysia specific acetyltransferase neuo
|
|
|
| 40 | c4n6bB_ |
|
not modelled |
99.9 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:serine acetyltransferase apoenzyme;
PDBTitle: soybean serine acetyltransferase complexed with coa
|
|
|
| 41 | d1xata_ |
|
not modelled |
99.9 |
16 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Galactoside acetyltransferase-like |
|
|
| 42 | c3ixcA_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:hexapeptide transferase family protein;
PDBTitle: crystal structure of hexapeptide transferase family protein from2 anaplasma phagocytophilum
|
|
|
| 43 | c3mc4A_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:ww/rsp5/wwp domain:bacterial transferase hexapeptide
PDBTitle: crystal structure of ww/rsp5/wwp domain: bacterial transferase2 hexapeptide repeat: serine o-acetyltransferase from brucella3 melitensis
|
|
|
| 44 | c4n27D_ |
|
not modelled |
99.9 |
22 |
PDB header:transferase
Chain: D: PDB Molecule:bacterial transferase hexapeptide repeat;
PDBTitle: x-ray structure of brucella abortus rica
|
|
|
| 45 | c4aa7A_ |
|
not modelled |
99.9 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: e.coli glmu in complex with an antibacterial inhibitor
|
|
|
| 46 | d2oi6a1 |
|
not modelled |
99.9 |
21 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:GlmU C-terminal domain-like |
|
|
| 47 | c3eg4A_ |
|
not modelled |
99.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:2,3,4,5-tetrahydropyridine-2,6-dicarboxylate n-
PDBTitle: crystal structure of 2,3,4,5-tetrahydropyridine-2-2 carboxylate n-succinyltransferase from brucella melitensis3 biovar abortus 2308
|
|
|
| 48 | c3d8vA_ |
|
not modelled |
99.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: crystal structure of glmu from mycobacterium tuberculosis2 in complex with uridine-diphosphate-n-acetylglucosamine
|
|
|
| 49 | c5ux9D_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: D: PDB Molecule:chloramphenicol acetyltransferase;
PDBTitle: the crystal structure of chloramphenicol acetyltransferase from vibrio2 fischeri es114
|
|
|
| 50 | c3foqA_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: crystal structure of n-acetylglucosamine-1-phosphate2 uridyltransferase (glmu) from mycobacterium tuberculosis in3 a cubic space group.
|
|
|
| 51 | c2wlgA_ |
|
not modelled |
99.9 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:polysialic acid o-acetyltransferase;
PDBTitle: crystallographic analysis of the polysialic acid o-2 acetyltransferase oatwy
|
|
|
| 52 | d1v3wa_ |
|
not modelled |
99.9 |
17 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:gamma-carbonic anhydrase-like |
|
|
| 53 | c3eevC_ |
|
not modelled |
99.9 |
16 |
PDB header:transferase
Chain: C: PDB Molecule:chloramphenicol acetyltransferase;
PDBTitle: crystal structure of chloramphenicol acetyltransferase vca0300 from2 vibrio cholerae o1 biovar eltor
|
|
|
| 54 | c3q1xA_ |
|
not modelled |
99.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:serine acetyltransferase;
PDBTitle: crystal structure of entamoeba histolytica serine acetyltransferase 12 in complex with l-serine
|
|
|
| 55 | c3r3rA_ |
|
not modelled |
99.9 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:ferripyochelin binding protein;
PDBTitle: structure of the yrda ferripyochelin binding protein from salmonella2 enterica
|
|
|
| 56 | d3tdta_ |
|
not modelled |
99.9 |
16 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:Tetrahydrodipicolinate-N-succinlytransferase, THDP-succinlytransferase, DapD |
|
|
| 57 | c3c8vA_ |
|
not modelled |
99.9 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:putative acetyltransferase;
PDBTitle: crystal structure of putative acetyltransferase (yp_390128.1) from2 desulfovibrio desulfuricans g20 at 2.28 a resolution
|
|
|
| 58 | c5afuU_ |
|
not modelled |
99.9 |
16 |
PDB header:motor protein
Chain: U: PDB Molecule:dynactin;
PDBTitle: cryo-em structure of dynein tail-dynactin-bicd2n complex
|
|
|
| 59 | c6iveA_ |
|
not modelled |
99.9 |
21 |
PDB header:metal binding protein
Chain: A: PDB Molecule:ferripyochelin-binding protein;
PDBTitle: molecular structure of a thermostable and a zinc ion binding gamma-2 class carbonic anhydrase
|
|
|
| 60 | c4mfgA_ |
|
not modelled |
99.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:putative acyltransferase;
PDBTitle: 2.0 angstrom resolution crystal structure of putative carbonic2 anhydrase from clostridium difficile.
|
|
|
| 61 | c1hm8A_ |
|
not modelled |
99.9 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine-1-phosphate uridyltransferase;
PDBTitle: crystal structure of s.pneumoniae n-acetylglucosamine-1-phosphate2 uridyltransferase, glmu, bound to acetyl coenzyme a
|
|
|
| 62 | c6cktA_ |
|
not modelled |
99.9 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:2,3,4,5-tetrahydropyridine-2,6-dicarboxylate n-
PDBTitle: crystal structure of 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate n-2 succinyltransferase from legionella pneumophila philadelphia 1
|
|
|
| 63 | d1xhda_ |
|
not modelled |
99.8 |
18 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:gamma-carbonic anhydrase-like |
|
|
| 64 | c3r1wA_ |
|
not modelled |
99.8 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:carbonic anhydrase;
PDBTitle: crystal structure of a carbonic anhydrase from a crude oil degrading2 psychrophilic library
|
|
|
| 65 | c5afuV_ |
|
not modelled |
99.8 |
14 |
PDB header:motor protein
Chain: V: PDB Molecule:dynactin;
PDBTitle: cryo-em structure of dynein tail-dynactin-bicd2n complex
|
|
|
| 66 | c3f1xA_ |
|
not modelled |
99.8 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:serine acetyltransferase;
PDBTitle: three dimensional structure of the serine acetyltransferase from2 bacteroides vulgatus, northeast structural genomics consortium target3 bvr62.
|
|
|
| 67 | c2v0hA_ |
|
not modelled |
99.8 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: characterization of substrate binding and catalysis of the2 potential antibacterial target n-acetylglucosamine-1-3 phosphate uridyltransferase (glmu)
|
|
|
| 68 | c3kwcD_ |
|
not modelled |
99.8 |
20 |
PDB header:lyase, protein binding, photosynthesis
Chain: D: PDB Molecule:carbon dioxide concentrating mechanism protein;
PDBTitle: oxidized, active structure of the beta-carboxysomal gamma-carbonic2 anhydrase, ccmm
|
|
|
| 69 | c2oi6A_ |
|
not modelled |
99.8 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: e. coli glmu- complex with udp-glcnac, coa and glcn-1-po4
|
|
|
| 70 | c3r5dA_ |
|
not modelled |
99.8 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:tetrahydrodipicolinate n-succinyletransferase;
PDBTitle: pseudomonas aeruginosa dapd (pa3666) apoprotein
|
|
|
| 71 | c5e3pA_ |
|
not modelled |
99.8 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:2,3,4,5-tetrahydropyridine-2,6-dicarboxylate n-
PDBTitle: crystal strcuture of dapd from corynebacterium glutamicum
|
|
|
| 72 | c3tv0A_ |
|
not modelled |
99.8 |
16 |
PDB header:structural protein
Chain: A: PDB Molecule:dynactin subunit 6;
PDBTitle: structure of dynactin p27 subunit
|
|
|
| 73 | c1qreA_ |
|
not modelled |
99.8 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:carbonic anhydrase;
PDBTitle: a closer look at the active site of gamma-carbonic anhydrases: high2 resolution crystallographic studies of the carbonic anhydrase from3 methanosarcina thermophila
|
|
|
| 74 | d1qrea_ |
|
not modelled |
99.8 |
20 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:gamma-carbonic anhydrase-like |
|
|
| 75 | c3fsyC_ |
|
not modelled |
99.8 |
17 |
PDB header:transferase
Chain: C: PDB Molecule:tetrahydrodipicolinate n-succinyltransferase;
PDBTitle: structure of tetrahydrodipicolinate n-succinyltransferase2 (rv1201c;dapd) in complex with succinyl-coa from mycobacterium3 tuberculosis
|
|
|
| 76 | c3kwdA_ |
|
not modelled |
99.7 |
19 |
PDB header:lyase, protein binding, photosynthesis
Chain: A: PDB Molecule:carbon dioxide concentrating mechanism protein;
PDBTitle: inactive truncation of the beta-carboxysomal gamma-carbonic anhydrase,2 ccmm, form 1
|
|
|
| 77 | c2ggqA_ |
|
not modelled |
99.7 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:401aa long hypothetical glucose-1-phosphate
PDBTitle: complex of hypothetical glucose-1-phosphate thymidylyltransferase from2 sulfolobus tokodaii
|
|
|
| 78 | c5vmkB_ |
|
not modelled |
99.7 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:bifunctional protein glmu;
PDBTitle: crystal structure of a bifunctional glmu udp-n-acetylglucosamine2 diphosphorylase/glucosamine-1- phosphate n-acetyltransferase from3 acinetobacter baumannii
|
|
|
| 79 | d2f9ca1 |
|
not modelled |
99.6 |
17 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:YdcK-like |
|
|
| 80 | c6i3mG_ |
|
not modelled |
99.6 |
20 |
PDB header:translation
Chain: G: PDB Molecule:translation initiation factor eif-2b subunit epsilon;
PDBTitle: eif2b:eif2 complex, phosphorylated on eif2 alpha serine 52.
|
|
|
| 81 | c5b04I_ |
|
not modelled |
99.6 |
13 |
PDB header:translation
Chain: I: PDB Molecule:probable translation initiation factor eif-2b subunit
PDBTitle: crystal structure of the eukaryotic translation initiation factor 2b2 from schizosaccharomyces pombe
|
|
|
| 82 | c6jlwJ_ |
|
not modelled |
99.5 |
7 |
PDB header:translation
Chain: J: PDB Molecule:translation initiation factor eif-2b subunit epsilon;
PDBTitle: eif2 - eif2b complex
|
|
|
| 83 | c6ezoJ_ |
|
not modelled |
99.5 |
13 |
PDB header:membrane protein
Chain: J: PDB Molecule:human eukaryotic initiation factor eif2b epsilon subunits;
PDBTitle: eukaryotic initiation factor eif2b in complex with isrib
|
|
|
| 84 | c5b04F_ |
|
not modelled |
99.5 |
16 |
PDB header:translation
Chain: F: PDB Molecule:probable translation initiation factor eif-2b subunit
PDBTitle: crystal structure of the eukaryotic translation initiation factor 2b2 from schizosaccharomyces pombe
|
|
|
| 85 | c3d98A_ |
|
not modelled |
99.4 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: crystal structure of glmu from mycobacterium tuberculosis, ligand-free2 form
|
|
|
| 86 | c2rijA_ |
|
not modelled |
99.4 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:putative 2,3,4,5-tetrahydropyridine-2-carboxylate n-
PDBTitle: crystal structure of a putative 2,3,4,5-tetrahydropyridine-2-2 carboxylate n-succinyltransferase (cj1605c, dapd) from campylobacter3 jejuni at 1.90 a resolution
|
|
|
| 87 | c6qg2F_ |
|
not modelled |
99.4 |
11 |
PDB header:translation
Chain: F: PDB Molecule:translation initiation factor eif-2b subunit gamma;
PDBTitle: structure of eif2b-eif2 (phosphorylated at ser51) complex (model a)
|
|
|
| 88 | c2qkxA_ |
|
not modelled |
99.3 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: n-acetyl glucosamine 1-phosphate uridyltransferase from mycobacterium2 tuberculosis complex with n-acetyl glucosamine 1-phosphate
|
|
|
| 89 | d1fxja1 |
|
not modelled |
99.2 |
20 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:GlmU C-terminal domain-like |
|
|
| 90 | d1yp2a1 |
|
not modelled |
99.2 |
11 |
Fold:Single-stranded left-handed beta-helix
Superfamily:Trimeric LpxA-like enzymes
Family:GlmU C-terminal domain-like |
|
|
| 91 | c6ezoF_ |
|
not modelled |
99.2 |
20 |
PDB header:membrane protein
Chain: F: PDB Molecule:translation initiation factor eif-2b subunit gamma;
PDBTitle: eukaryotic initiation factor eif2b in complex with isrib
|
|
|
| 92 | c1yp3C_ |
|
not modelled |
99.2 |
11 |
PDB header:transferase
Chain: C: PDB Molecule:glucose-1-phosphate adenylyltransferase small
PDBTitle: crystal structure of potato tuber adp-glucose2 pyrophosphorylase in complex with atp
|
|
|
| 93 | c5l6sF_ |
|
not modelled |
99.1 |
13 |
PDB header:transferase
Chain: F: PDB Molecule:glucose-1-phosphate adenylyltransferase;
PDBTitle: crystal structure of e. coli adp-glucose pyrophosphorylase (agpase) in2 complex with a positive allosteric regulator beta-fructose-1,6-3 diphosphate (fbp) - agpase*fbp
|
|
|
| 94 | c1fwyA_ |
|
not modelled |
98.9 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine pyrophosphorylase;
PDBTitle: crystal structure of n-acetylglucosamine 1-phosphate2 uridyltransferase bound to udp-glcnac
|
|
|
| 95 | c3brkX_ |
|
not modelled |
98.9 |
16 |
PDB header:transferase
Chain: X: PDB Molecule:glucose-1-phosphate adenylyltransferase;
PDBTitle: crystal structure of adp-glucose pyrophosphorylase from agrobacterium2 tumefaciens
|
|
|
| 96 | c3nklA_ |
|
not modelled |
98.7 |
14 |
PDB header:oxidoreductase/lyase
Chain: A: PDB Molecule:udp-d-quinovosamine 4-dehydrogenase;
PDBTitle: crystal structure of udp-d-quinovosamine 4-dehydrogenase from vibrio2 fischeri
|
|
|
| 97 | c2dt5A_ |
|
not modelled |
98.2 |
15 |
PDB header:dna binding protein
Chain: A: PDB Molecule:at-rich dna-binding protein;
PDBTitle: crystal structure of ttha1657 (at-rich dna-binding protein) from2 thermus thermophilus hb8
|
|
|
| 98 | d2py6a1 |
|
not modelled |
98.0 |
17 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:FkbM-like |
|
|
| 99 | c2vt2A_ |
|
not modelled |
97.9 |
6 |
PDB header:transcription
Chain: A: PDB Molecule:redox-sensing transcriptional repressor rex;
PDBTitle: structure and functional properties of the bacillus subtilis2 transcriptional repressor rex
|
|
|
| 100 | c3wg9D_ |
|
not modelled |
97.8 |
9 |
PDB header:transcription
Chain: D: PDB Molecule:redox-sensing transcriptional repressor rex;
PDBTitle: crystal structure of rsp, a rex-family repressor
|
|
|
| 101 | d2dt5a2 |
|
not modelled |
97.5 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Transcriptional repressor Rex, C-terminal domain |
|
|
| 102 | c5zz5D_ |
|
not modelled |
97.4 |
14 |
PDB header:gene regulation
Chain: D: PDB Molecule:redox-sensing transcriptional repressor rex;
PDBTitle: redox-sensing transcriptional repressor rex
|
|
|
| 103 | c3ketA_ |
|
not modelled |
97.2 |
12 |
PDB header:transcription/dna
Chain: A: PDB Molecule:redox-sensing transcriptional repressor rex;
PDBTitle: crystal structure of a rex-family transcriptional regulatory protein2 from streptococcus agalactiae bound to a palindromic operator
|
|
|
| 104 | c3gfgB_ |
|
not modelled |
95.0 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:uncharacterized oxidoreductase yvaa;
PDBTitle: structure of putative oxidoreductase yvaa from bacillus subtilis in2 triclinic form
|
|
|
| 105 | d1kjqa2 |
|
not modelled |
94.8 |
14 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
|
|
| 106 | d1ek6a_ |
|
not modelled |
94.8 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 107 | c6norB_ |
|
not modelled |
94.5 |
9 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative nad dependent dehydrogenase;
PDBTitle: crystal structure of gend2 from gentamicin a biosynthesis in complex2 with nad
|
|
|
| 108 | c3euwB_ |
|
not modelled |
94.4 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:myo-inositol dehydrogenase;
PDBTitle: crystal structure of a myo-inositol dehydrogenase from corynebacterium2 glutamicum atcc 13032
|
|
|
| 109 | d1f06a1 |
|
not modelled |
94.4 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 110 | c3dapB_ |
|
not modelled |
94.2 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:diaminopimelic acid dehydrogenase;
PDBTitle: c. glutamicum dap dehydrogenase in complex with nadp+ and2 the inhibitor 5s-isoxazoline
|
|
|
| 111 | c2hjrK_ |
|
not modelled |
94.1 |
18 |
PDB header:oxidoreductase
Chain: K: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of cryptosporidium parvum malate2 dehydrogenase
|
|
|
| 112 | c3dhnA_ |
|
not modelled |
93.9 |
16 |
PDB header:isomerase, lyase
Chain: A: PDB Molecule:nad-dependent epimerase/dehydratase;
PDBTitle: crystal structure of the putative epimerase q89z24_bactn from2 bacteroides thetaiotaomicron. northeast structural genomics3 consortium target btr310.
|
|
|
| 113 | c4wpgA_ |
|
not modelled |
93.8 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dtdp-4-dehydrorhamnose reductase;
PDBTitle: group a streptococcus gaca is an essential dtdp-4-dehydrorhamnose2 reductase (rmld)
|
|
|
| 114 | c3k96B_ |
|
not modelled |
93.8 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glycerol-3-phosphate dehydrogenase [nad(p)+];
PDBTitle: 2.1 angstrom resolution crystal structure of glycerol-3-phosphate2 dehydrogenase (gpsa) from coxiella burnetii
|
|
|
| 115 | c3e18A_ |
|
not modelled |
93.7 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of nad-binding protein from listeria innocua
|
|
|
| 116 | c3sc6F_ |
|
not modelled |
93.6 |
23 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:dtdp-4-dehydrorhamnose reductase;
PDBTitle: 2.65 angstrom resolution crystal structure of dtdp-4-dehydrorhamnose2 reductase (rfbd) from bacillus anthracis str. ames in complex with3 nadp
|
|
|
| 117 | c2ph5A_ |
|
not modelled |
93.6 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:homospermidine synthase;
PDBTitle: crystal structure of the homospermidine synthase hss from legionella2 pneumophila in complex with nad, northeast structural genomics target3 lgr54
|
|
|
| 118 | c2gr2A_ |
|
not modelled |
93.6 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ferredoxin reductase;
PDBTitle: crystal structure of ferredoxin reductase, bpha4 (oxidized form)
|
|
|
| 119 | c5gmoA_ |
|
not modelled |
93.3 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein induced by osmotic stress;
PDBTitle: x-ray structure of carbonyl reductase sscr
|
|
|
| 120 | c3m2tA_ |
|
not modelled |
93.1 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable dehydrogenase;
PDBTitle: the crystal structure of dehydrogenase from chromobacterium2 violaceum
|
|
|