1 c5fceB_
33.7
31
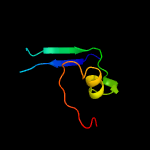
PDB header: cell adhesionChain: B: PDB Molecule: lpxtg family cell surface protein fms2;PDBTitle: the crystal structure of the ligand binding region of serine-glutamate2 repeat protein a (sgra) of enterococcus faecium
2 c4ffbC_
29.7
8
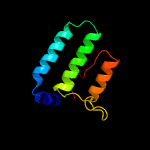
PDB header: hydrolaseChain: C: PDB Molecule: protein stu2;PDBTitle: a tog:alpha/beta-tubulin complex structure reveals conformation-based2 mechanisms for a microtubule polymerase
3 c4j7hA_
28.7
28
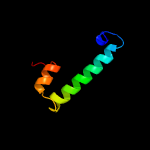
PDB header: biosynthetic proteinChain: A: PDB Molecule: evaa 2,3-dehydratase;PDBTitle: crystal structure of evaa, a 2,3-dehydratase in complex with dtdp-2 benzene and dtdp-rhamnose
4 c3f6wE_
24.4
7
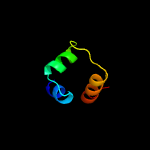
PDB header: dna binding proteinChain: E: PDB Molecule: xre-family like protein;PDBTitle: xre-family like protein from pseudomonas syringae pv. tomato str.2 dc3000
5 c3eusB_
23.7
5
PDB header: dna binding proteinChain: B: PDB Molecule: dna-binding protein;PDBTitle: the crystal structure of the dna binding protein from silicibacter2 pomeroyi
6 c1m6vE_
23.4
14
PDB header: ligaseChain: E: PDB Molecule: carbamoyl phosphate synthetase large chain;PDBTitle: crystal structure of the g359f (small subunit) point mutant of2 carbamoyl phosphate synthetase
7 d1a9xa4
21.5
10
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like
8 d2bs2b2
20.2
19
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins
9 d1y7ya1
19.2
5
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: SinR domain-like
10 d1kkha2
18.5
14
Fold: Ferredoxin-likeSuperfamily: GHMP Kinase, C-terminal domainFamily: Mevalonate kinase
11 c3p9aI_
18.1
13
PDB header: dna binding proteinChain: I: PDB Molecule: dna-packaging protein gp3;PDBTitle: an atomic view of the nonameric small terminase subunit of2 bacteriophage p22
12 d1llib_
16.2
10
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors
13 d1td6a_
14.5
15
Fold: Hypothetical protein MPN330Superfamily: Hypothetical protein MPN330Family: Hypothetical protein MPN330
14 c3jb9c_
14.4
9
PDB header: rna binding protein/rnaChain: C: PDB Molecule: u5 snrna;PDBTitle: cryo-em structure of the yeast spliceosome at 3.6 angstrom resolution
15 c2jvlA_
14.4
10
PDB header: transcriptionChain: A: PDB Molecule: trmbf1;PDBTitle: nmr structure of the c-terminal domain of mbf1 of trichoderma reesei
16 d2cpwa1
14.2
33
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain
17 c3trbA_
13.7
14
PDB header: dna binding proteinChain: A: PDB Molecule: virulence-associated protein i;PDBTitle: structure of an addiction module antidote protein of a higa (higa)2 family from coxiella burnetii
18 c5fdkD_
13.6
13
PDB header: hydrolaseChain: D: PDB Molecule: holliday junction resolvase recu;PDBTitle: crystal structure of recu(d88n) in complex with palindromic dna duplex
19 d1rioa_
12.7
7
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors
20 c3omtA_
12.5
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: putative antitoxin component, chu_2935 protein, from xre family from2 prevotella buccae.
21 c5lxlA_
not modelled
12.5
21
PDB header: viral proteinChain: A: PDB Molecule: decoration protein;PDBTitle: nmr structure of the n-terminal domain of the bacteriophage t52 decoration protein pb10
22 c3f52A_
not modelled
12.4
17
PDB header: transcription activatorChain: A: PDB Molecule: clp gene regulator (clgr);PDBTitle: crystal structure of the clp gene regulator clgr from c. glutamicum
23 d1kf6b2
not modelled
12.1
16
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins
24 c2yiaE_
not modelled
11.7
16
PDB header: transferaseChain: E: PDB Molecule: rna-directed rna polymerase;PDBTitle: structure of the rna polymerase vp1 from infectious pancreatic2 necrosis virus
25 c2yibC_
not modelled
11.5
16
PDB header: transferaseChain: C: PDB Molecule: rna-directed rna polymerase;PDBTitle: structure of the rna polymerase vp1 from infectious pancreatic2 necrosis virus
26 c2kpjA_
not modelled
11.3
17
PDB header: transcription regulatorChain: A: PDB Molecule: sos-response transcriptional repressor, lexa;PDBTitle: solution structure of protein sos-response transcriptional2 repressor, lexa from eubacterium rectale. northeast3 structural genomics consortium target err9a
27 c2y27B_
not modelled
11.3
15
PDB header: ligaseChain: B: PDB Molecule: phenylacetate-coenzyme a ligase;PDBTitle: crystal structure of paak1 in complex with atp from burkholderia2 cenocepacia
28 c3mlfC_
not modelled
10.6
10
PDB header: transcription regulatorChain: C: PDB Molecule: transcriptional regulator;PDBTitle: putative transcriptional regulator from staphylococcus aureus.
29 d1utxa_
not modelled
10.3
7
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: SinR domain-like
30 c2yibD_
not modelled
10.3
16
PDB header: transferaseChain: D: PDB Molecule: rna-directed rna polymerase;PDBTitle: structure of the rna polymerase vp1 from infectious pancreatic2 necrosis virus
31 c2ls01_
not modelled
10.0
33
PDB header: hydrolaseChain: 1: PDB Molecule: zoocin a endopeptidase;PDB Fragment: unp residues 170-285;
PDBTitle: solution structure of the target recognition domain of zoocin a
32 d1e3ha4
not modelled
9.9
15
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Prokaryotic type KH domain (KH-domain type II)Family: Prokaryotic type KH domain (KH-domain type II)
33 c1y6uA_
not modelled
9.7
14
PDB header: dna binding proteinChain: A: PDB Molecule: excisionase from transposon tn916;PDBTitle: the structure of the excisionase (xis) protein from2 conjugative transposon tn916 provides insights into the3 regulation of heterobivalent tyrosine recombinases
34 d3bexa1
not modelled
9.7
11
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: CoaX-like
35 c3ivpD_
not modelled
9.2
10
PDB header: dna binding proteinChain: D: PDB Molecule: putative transposon-related dna-binding protein;PDBTitle: the structure of a possible transposon-related dna-binding protein2 from clostridium difficile 630.
36 c2ewtA_
not modelled
9.0
18
PDB header: dna binding proteinChain: A: PDB Molecule: putative dna-binding protein;PDBTitle: crystal structure of the dna-binding domain of bldd
37 d1jqoa_
not modelled
9.0
20
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Phosphoenolpyruvate carboxylase
38 c1jqoA_
not modelled
9.0
20
PDB header: lyaseChain: A: PDB Molecule: phosphoenolpyruvate carboxylase;PDBTitle: crystal structure of c4-form phosphoenolpyruvate carboxylase from2 maize
39 c1y9qA_
not modelled
8.6
15
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, hth_3 family;PDBTitle: crystal structure of hth_3 family transcriptional regulator2 from vibrio cholerae
40 c3noeA_
not modelled
8.5
30
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from pseudomonas2 aeruginosa
41 c3bs3A_
not modelled
8.4
10
PDB header: dna binding proteinChain: A: PDB Molecule: putative dna-binding protein;PDBTitle: crystal structure of a putative dna-binding protein from bacteroides2 fragilis
42 c2cpwA_
not modelled
8.3
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: cbl-interacting protein sts-1 variant;PDBTitle: solution structure of rsgi ruh-031, a uba domain from human2 cdna
43 c4wbdA_
not modelled
8.3
13
PDB header: ligaseChain: A: PDB Molecule: bshc;PDBTitle: the crystal structure of bshc from bacillus subtilis complexed with2 citrate and adp
44 c1rznB_
not modelled
8.3
14
PDB header: dna binding proteinChain: B: PDB Molecule: recombination protein u;PDBTitle: crystal structure of penicillin-binding protein-related2 factor a from bacillus subtilis.
45 c1k47F_
not modelled
8.1
13
PDB header: transferaseChain: F: PDB Molecule: phosphomevalonate kinase;PDBTitle: crystal structure of the streptococcus pneumoniae2 phosphomevalonate kinase (pmk)
46 c3iz5t_
not modelled
8.1
25
PDB header: ribosomeChain: T: PDB Molecule: 60s ribosomal protein l19 (l19e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
47 d1b0na2
not modelled
8.0
10
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: SinR domain-like
48 c3clcC_
not modelled
7.9
2
PDB header: transcription regulator/dnaChain: C: PDB Molecule: regulatory protein;PDBTitle: crystal structure of the restriction-modification controller protein2 c.esp1396i tetramer in complex with its natural 35 base-pair operator
49 c3dnvB_
not modelled
7.8
7
PDB header: transcription/dnaChain: B: PDB Molecule: hth-type transcriptional regulator hipb;PDBTitle: mdt protein
50 c6f8sA_
not modelled
7.7
20
PDB header: toxinChain: A: PDB Molecule: xre family transcriptional regulator;PDBTitle: toxin-antitoxin complex grata
51 d1adra_
not modelled
7.6
17
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors
52 c1y1oC_
not modelled
7.6
11
PDB header: recombinationChain: C: PDB Molecule: penicillin-binding protein-related factor a;PDBTitle: x-ray crystal structure of penicillin-binding protein-2 related factor a from bacillus stearothermophilus
53 d1y1oa_
not modelled
7.6
11
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: RecU-like
54 d1x57a1
not modelled
7.5
5
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: EDF1-like
55 d2b5aa1
not modelled
7.5
5
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: SinR domain-like
56 c3vk0B_
not modelled
7.4
2
PDB header: dna binding proteinChain: B: PDB Molecule: transcriptional regulator;PDBTitle: crystal structure of hypothetical transcription factor nhtf from2 neisseria
57 d1rzna_
not modelled
7.4
14
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: RecU-like
58 c3skqA_
not modelled
7.3
20
PDB header: metal transportChain: A: PDB Molecule: mitochondrial distribution and morphology protein 38;PDBTitle: mdm38 is a 14-3-3-like receptor and associates with the protein2 synthesis machinery at the inner mitochondrial membrane
59 d2gx8a1
not modelled
7.1
15
Fold: NIF3 (NGG1p interacting factor 3)-likeSuperfamily: NIF3 (NGG1p interacting factor 3)-likeFamily: NIF3 (NGG1p interacting factor 3)-like
60 c3t76A_
not modelled
7.1
10
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator vanug;PDBTitle: crystal structure of transcriptional regulator vanug, form ii
61 c6h56A_
not modelled
7.0
11
PDB header: metal binding proteinChain: A: PDB Molecule: effector domain of pseudomonas aeruginosa vgrg2b;PDBTitle: effector domain of pseudomonas aeruginosa vgrg2b
62 c3op9A_
not modelled
6.9
17
PDB header: transcription regulatorChain: A: PDB Molecule: pli0006 protein;PDBTitle: crystal structure of transcriptional regulator from listeria innocua
63 c4mcxE_
not modelled
6.9
5
PDB header: toxinChain: E: PDB Molecule: antidote protein;PDBTitle: p. vulgaris higba structure, crystal form 2
64 c1oheA_
not modelled
6.8
14
PDB header: hydrolaseChain: A: PDB Molecule: cdc14b2 phosphatase;PDBTitle: structure of cdc14b phosphatase with a peptide ligand
65 c6bi7B_
not modelled
6.8
21
PDB header: replicationChain: B: PDB Molecule: dna polymerase zeta catalytic subunit;PDBTitle: crystal structure of rev7-wt/rev3 as a monomer under high-salt2 conditions
66 d1edza2
not modelled
6.8
21
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Tetrahydrofolate dehydrogenase/cyclohydrolase
67 c6mghB_
not modelled
6.7
9
PDB header: fluorescent proteinChain: B: PDB Molecule: mirfp670nano;PDBTitle: x-ray structure of monomeric near-infrared fluorescent protein2 mirfp670nano
68 c5uk3J_
not modelled
6.3
7
PDB header: lyaseChain: J: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of cyanase from t. urticae
69 c3rnlA_
not modelled
6.1
17
PDB header: transferaseChain: A: PDB Molecule: sulfotransferase;PDBTitle: crystal structure of sulfotransferase from alicyclobacillus2 acidocaldarius
70 c2mezA_
not modelled
6.0
7
PDB header: rna binding proteinChain: A: PDB Molecule: multiprotein bridging factor (mbp-like);PDBTitle: flexible anchoring of archaeal mbf1 on ribosomes suggests role as2 recruitment factor
71 d1sq8a_
not modelled
5.9
10
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors
72 c3n2xB_
not modelled
5.8
22
PDB header: lyaseChain: B: PDB Molecule: uncharacterized protein yage;PDBTitle: crystal structure of yage, a prophage protein belonging to the2 dihydrodipicolinic acid synthase family from e. coli k12 in complex3 with pyruvate
73 d2r1jl1
not modelled
5.8
17
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors
74 c2yxgD_
not modelled
5.8
21
PDB header: lyaseChain: D: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihyrodipicolinate synthase (dapa)
75 c2o38A_
not modelled
5.8
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: putative xre family transcriptional regulator
76 d2o38a1
not modelled
5.8
22
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: NE1354
77 d2a6ca1
not modelled
5.7
9
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: NE1354
78 c2xxzA_
not modelled
5.7
10
PDB header: oxidoreductaseChain: A: PDB Molecule: lysine-specific demethylase 6b;PDBTitle: crystal structure of the human jmjd3 jumonji domain
79 c2gx8B_
not modelled
5.6
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: nif3-related protein;PDBTitle: the crystal structure of bacillus cereus protein related to nif3
80 c4xd7G_
not modelled
5.5
13
PDB header: hydrolaseChain: G: PDB Molecule: atp synthase gamma chain;PDBTitle: structure of thermophilic f1-atpase inhibited by epsilon subunit
81 c3izbQ_
not modelled
5.5
32
PDB header: ribosomeChain: Q: PDB Molecule: 40s ribosomal protein rps17 (s17e);PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
82 c4b0sA_
not modelled
5.3
22
PDB header: hydrolaseChain: A: PDB Molecule: deamidase-depupylase dop;PDBTitle: structure of the deamidase-depupylase dop of the prokaryotic2 ubiquitin-like modification pathway in complex with atp
83 c2ef8A_
not modelled
5.2
15
PDB header: transcription regulatorChain: A: PDB Molecule: putative transcription factor;PDBTitle: crystal structure of c.ecot38is
84 c3bdnB_
not modelled
5.2
8
PDB header: transcription/dnaChain: B: PDB Molecule: lambda repressor;PDBTitle: crystal structure of the lambda repressor
85 c3eb2A_
not modelled
5.2
29
PDB header: lyaseChain: A: PDB Molecule: putative dihydrodipicolinate synthetase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
86 c5ocdD_
not modelled
5.1
10
PDB header: rna binding proteinChain: D: PDB Molecule: cyclodipeptide synthase;PDBTitle: structure of a cdps from fluoribacter dumoffii
87 c6bi7D_
not modelled
5.1
22
PDB header: replicationChain: D: PDB Molecule: dna polymerase zeta catalytic subunit;PDBTitle: crystal structure of rev7-wt/rev3 as a monomer under high-salt2 conditions
88 c3j3aR_
not modelled
5.1
21
PDB header: ribosomeChain: R: PDB Molecule: 40s ribosomal protein s17;PDBTitle: structure of the human 40s ribosomal proteins