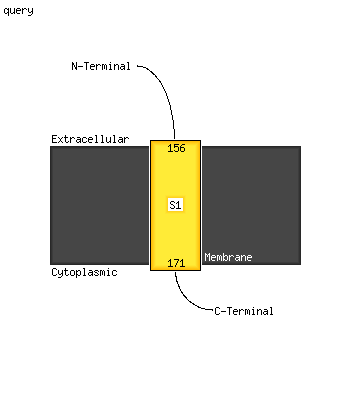
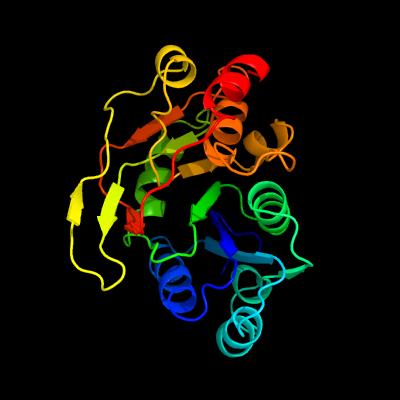
| 1 | c2z86D_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: D: PDB Molecule:chondroitin synthase;
PDBTitle: crystal structure of chondroitin polymerase from escherichia coli2 strain k4 (k4cp) complexed with udp-glcua and udp
|
|
|
|
| 2 | d1xhba2
|
|
 |
100.0 |
15 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Polypeptide N-acetylgalactosaminyltransferase 1, N-terminal domain |
|
|
|
| 3 | c1xhbA_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:polypeptide n-acetylgalactosaminyltransferase 1;
PDBTitle: the crystal structure of udp-galnac: polypeptide alpha-n-2 acetylgalactosaminyltransferase-t1
|
|
|
|
| 4 | c6h4mA_
|
|
 |
100.0 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:probable ss-1,3-n-acetylglucosaminyltransferase;
PDBTitle: tarp-udp-glcnac-3rbop
|
|
|
|
| 5 | c6e4rB_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:polypeptide n-acetylgalactosaminyltransferase 9;
PDBTitle: crystal structure of the drosophila melanogaster polypeptide n-2 acetylgalactosaminyl transferase pgant9b
|
|
|
|
| 6 | c2d7iA_
|
|
 |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:polypeptide n-acetylgalactosaminyltransferase 10;
PDBTitle: crystal structure of pp-galnac-t10 with udp, galnac and mn2+
|
|
|
|
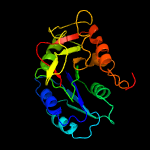
| 7 | c5tz8C_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: C: PDB Molecule:glycosyl transferase;
PDBTitle: crystal structure of s. aureus tars
|
|
|
|
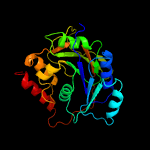
| 8 | c6iwqE_
|
|
 |
100.0 |
13 |
PDB header:transferase
Chain: E: PDB Molecule:n-acetylgalactosaminyltransferase 7;
PDBTitle: crystal structure of galnac-t7 with mn2+
|
|
|
|
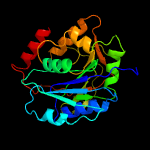
| 9 | c2ffuA_
|
|
 |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:polypeptide n-acetylgalactosaminyltransferase 2;
PDBTitle: crystal structure of human ppgalnact-2 complexed with udp and ea2
|
|
|
|
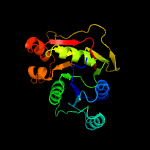
| 10 | c5nqaA_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:polypeptide n-acetylgalactosaminyltransferase 4;
PDBTitle: crystal structure of galnac-t4 in complex with the monoglycopeptide 3
|
|
|
|
| 11 | c4hg6A_
|
|
 |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:cellulose synthase subunit a;
PDBTitle: structure of a cellulose synthase - cellulose translocation2 intermediate
|
|
|
|
| 12 | c5mm1A_
|
|
 |
100.0 |
15 |
PDB header:membrane protein
Chain: A: PDB Molecule:dolichol monophosphate mannose synthase;
PDBTitle: dolichyl phosphate mannose synthase in complex with gdp and dolichyl2 phosphate mannose
|
|
|
|
| 13 | d1qg8a_
|
|
 |
100.0 |
20 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Spore coat polysaccharide biosynthesis protein SpsA |
|
|
|
| 14 | c5heaA_
|
|
 |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:putative glycosyltransferase (galt1);
PDBTitle: cgt structure in hexamer
|
|
|
|
| 15 | c3f1yC_
|
|
 |
100.0 |
16 |
PDB header:transferase
Chain: C: PDB Molecule:mannosyl-3-phosphoglycerate synthase;
PDBTitle: mannosyl-3-phosphoglycerate synthase from rubrobacter xylanophilus
|
|
|
|
| 16 | c3ckvA_
|
|
 |
100.0 |
14 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of a mycobacterial protein
|
|
|
|
| 17 | c3bcvA_
|
|
 |
99.9 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:putative glycosyltransferase protein;
PDBTitle: crystal structure of a putative glycosyltransferase from bacteroides2 fragilis
|
|
|
|
| 18 | c4fixA_
|
|
 |
99.9 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:udp-galactofuranosyl transferase glft2;
PDBTitle: crystal structure of glft2
|
|
|
|
| 19 | c1omxB_
|
|
 |
99.9 |
9 |
PDB header:transferase
Chain: B: PDB Molecule:alpha-1,4-n-acetylhexosaminyltransferase extl2;
PDBTitle: crystal structure of mouse alpha-1,4-n-acetylhexosaminyltransferase2 (extl2)
|
|
|
|
| 20 | d1omza_
|
|
 |
99.9 |
9 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Exostosin |
|
|
|
| 21 | c6p61D_ |
|
not modelled |
99.9 |
21 |
PDB header:transferase
Chain: D: PDB Molecule:glycosyltransferase;
PDBTitle: structure of a glycosyltransferase from leptospira borgpetersenii2 serovar hardjo-bovis (strain jb197)
|
|
|
| 22 | c5ekeB_ |
|
not modelled |
99.9 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:uncharacterized glycosyltransferase sll0501;
PDBTitle: structure of the polyisoprenyl-phosphate glycosyltransferase gtrb2 (f215a mutant)
|
|
|
| 23 | c3zf8A_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:mannan polymerase complexes subunit mnn9;
PDBTitle: crystal structure of saccharomyces cerevisiae mnn9 in2 complex with gdp and mn.
|
|
|
| 24 | c2qgiA_ |
|
not modelled |
99.8 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:atp synthase subunits region orf 6;
PDBTitle: the udp complex structure of the sixth gene product of the f1-atpase2 operon of rhodobacter blasticus
|
|
|
| 25 | c5ggfC_ |
|
not modelled |
99.8 |
16 |
PDB header:transferase, sugar binding protein
Chain: C: PDB Molecule:protein o-linked-mannose beta-1,2-n-
PDBTitle: crystal structure of human protein o-mannose beta-1,2-n-2 acetylglucosaminyltransferase form ii
|
|
|
| 26 | c5z8bB_ |
|
not modelled |
99.8 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:kfia protein;
PDBTitle: truncated n-acetylglucosaminyl transferase kfia from e. coli k5 strain2 apo form
|
|
|
| 27 | d1fo8a_ |
|
not modelled |
99.3 |
14 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:N-acetylglucosaminyltransferase I |
|
|
| 28 | c4irqB_ |
|
not modelled |
99.1 |
9 |
PDB header:transferase
Chain: B: PDB Molecule:beta-1,4-galactosyltransferase 7;
PDBTitle: crystal structure of catalytic domain of human beta1,2 4galactosyltransferase 7 in closed conformation in complex with3 manganese and udp
|
|
|
| 29 | d2bo4a1 |
|
not modelled |
99.1 |
14 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:MGS-like |
|
|
| 30 | c6fxyA_ |
|
not modelled |
98.6 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:procollagen-lysine,2-oxoglutarate 5-dioxygenase 3;
PDBTitle: crystal structure of full-length human lysyl hydroxylase lh3 -2 cocrystal with fe2+, mn2+, udp-gal - structure from long-wavelength3 s-sad
|
|
|
| 31 | d1pzta_ |
|
not modelled |
98.6 |
12 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:beta 1,4 galactosyltransferase (b4GalT1) |
|
|
| 32 | c5vcmA_ |
|
not modelled |
98.5 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:alpha-1,6-mannosyl-glycoprotein 2-beta-n-
PDBTitle: alpha-1,6-mannosyl-glycoprotein 2-beta-n-acetylglucosaminyltransferase2 with bound udp and manganese
|
|
|
| 33 | c3lw6A_ |
|
not modelled |
97.9 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:beta-4-galactosyltransferase 7;
PDBTitle: crystal structure of drosophila beta1,4-galactosyltransferase-7
|
|
|
| 34 | c4kt7A_ |
|
not modelled |
97.5 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: the crystal structure of 4-diphosphocytidyl-2c-methyl-d-2 erythritolsynthase from anaerococcus prevotii dsm 20548
|
|
|
| 35 | c2wvmA_ |
|
not modelled |
97.3 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:mannosyl-3-phosphoglycerate synthase;
PDBTitle: h309a mutant of mannosyl-3-phosphoglycerate synthase from2 thermus thermophilus hb27 in complex with3 gdp-alpha-d-mannose and mg(ii)
|
|
|
| 36 | c2zu8A_ |
|
not modelled |
96.7 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:mannosyl-3-phosphoglycerate synthase;
PDBTitle: crystal structure of mannosyl-3-phosphoglycerate synthase2 from pyrococcus horikoshii
|
|
|
| 37 | d1vh3a_ |
|
not modelled |
96.7 |
14 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 38 | c5vcsB_ |
|
not modelled |
96.7 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:alpha-1,6-mannosyl-glycoprotein 2-beta-n-
PDBTitle: alpha-1,6-mannosyl-glycoprotein 2-beta-n-acetylglucosaminyltransferase2 with bound acceptor
|
|
|
| 39 | c2d0jD_ |
|
not modelled |
96.3 |
11 |
PDB header:transferase
Chain: D: PDB Molecule:galactosylgalactosylxylosylprotein 3-beta-
PDBTitle: crystal structure of human glcat-s apo form
|
|
|
| 40 | c2wawA_ |
|
not modelled |
95.8 |
9 |
PDB header:unknown function
Chain: A: PDB Molecule:moba relate protein;
PDBTitle: crystal structure of mycobacterium tuberculosis rv0371c2 homolog from mycobacterium sp. strain jc1
|
|
|
| 41 | c5gvvF_ |
|
not modelled |
95.7 |
9 |
PDB header:transferase
Chain: F: PDB Molecule:glycosyl transferase family 8;
PDBTitle: crystal structure of the glycosyltransferase glye in streptococcus2 pneumoniae tigr4
|
|
|
| 42 | d3cu0a1 |
|
not modelled |
95.6 |
13 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:1,3-glucuronyltransferase |
|
|
| 43 | c3tztB_ |
|
not modelled |
95.6 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:glycosyl transferase family 8;
PDBTitle: the structure of a protein in glycosyl transferase family 8 from2 anaerococcus prevotii.
|
|
|
| 44 | c2px7A_ |
|
not modelled |
95.6 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: crystal structure of 2-c-methyl-d-erythritol 4-phosphate2 cytidylyltransferase from thermus thermophilus hb8
|
|
|
| 45 | d1v82a_ |
|
not modelled |
95.5 |
17 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:1,3-glucuronyltransferase |
|
|
| 46 | c6oewB_ |
|
not modelled |
95.5 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:cytidylyltransferase;
PDBTitle: structure of a cytidylyltransferase from leptospira borgpetersenii2 serovar hardjo-bovis (strain jb197)
|
|
|
| 47 | d1w77a1 |
|
not modelled |
95.4 |
14 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 48 | c4ys8B_ |
|
not modelled |
95.0 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: crystal structure of 2-c-methyl-d-erythritol 4-phosphate2 cytidylyltransferase (ispd) from burkholderia thailandensis
|
|
|
| 49 | d1mc3a_ |
|
not modelled |
94.5 |
10 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
|
|
| 50 | c4xwiA_ |
|
not modelled |
94.5 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:3-deoxy-manno-octulosonate cytidylyltransferase;
PDBTitle: x-ray crystal structure of cmp-kdo synthase from pseudomonas2 aeruginosa
|
|
|
| 51 | c5ddtA_ |
|
not modelled |
93.6 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: crystal structure of ispd from bacillus subtilis at 1.80 angstroms2 resolution, crystal form i
|
|
|
| 52 | c2xwlB_ |
|
not modelled |
93.3 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: crystal structure of ispd from mycobacterium smegmatis in complex with2 ctp and mg
|
|
|
| 53 | d1fxoa_ |
|
not modelled |
93.1 |
9 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
|
|
| 54 | c3okrA_ |
|
not modelled |
93.1 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: structure of mtb apo 2-c-methyl-d-erythritol 4-phosphate2 cytidyltransferase (ispd)
|
|
|
| 55 | d1lvwa_ |
|
not modelled |
93.1 |
10 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
|
|
| 56 | c3d5nB_ |
|
not modelled |
92.6 |
13 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:q97w15_sulso;
PDBTitle: crystal structure of the q97w15_sulso protein from sulfolobus2 solfataricus. nesg target ssr125.
|
|
|
| 57 | d1qwja_ |
|
not modelled |
92.6 |
13 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 58 | d1h5ra_ |
|
not modelled |
92.6 |
9 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
|
|
| 59 | c3foqA_ |
|
not modelled |
91.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: crystal structure of n-acetylglucosamine-1-phosphate2 uridyltransferase (glmu) from mycobacterium tuberculosis in3 a cubic space group.
|
|
|
| 60 | c3d8vA_ |
|
not modelled |
91.7 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: crystal structure of glmu from mycobacterium tuberculosis2 in complex with uridine-diphosphate-n-acetylglucosamine
|
|
|
| 61 | c6b5kA_ |
|
not modelled |
91.5 |
7 |
PDB header:transferase
Chain: A: PDB Molecule:glucose-1-phosphate thymidylyltransferase;
PDBTitle: mycobacterium tuberculosis rmla in complex with mg/dttp
|
|
|
| 62 | c2e8bA_ |
|
not modelled |
91.4 |
14 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:probable molybdopterin-guanine dinucleotide biosynthesis
PDBTitle: crystal structure of the putative protein (aq1419) from aquifex2 aeolicus vf5
|
|
|
| 63 | c4cvhA_ |
|
not modelled |
91.3 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:isoprenoid synthase domain-containing protein;
PDBTitle: crystal structure of human isoprenoid synthase domain-containing2 protein
|
|
|
| 64 | c1ga8A_ |
|
not modelled |
91.1 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:galactosyl transferase lgtc;
PDBTitle: crystal structure of galacosyltransferase lgtc in complex2 with donor and acceptor sugar analogs.
|
|
|
| 65 | d1ga8a_ |
|
not modelled |
91.1 |
16 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Galactosyltransferase LgtC |
|
|
| 66 | c2we9A_ |
|
not modelled |
91.0 |
9 |
PDB header:unknown function
Chain: A: PDB Molecule:moba-related protein;
PDBTitle: crystal structure of rv0371c from mycobacterium2 tuberculosis h37rv
|
|
|
| 67 | c3oamD_ |
|
not modelled |
90.5 |
13 |
PDB header:transferase
Chain: D: PDB Molecule:3-deoxy-manno-octulosonate cytidylyltransferase;
PDBTitle: crystal structure of cytidylyltransferase from vibrio cholerae
|
|
|
| 68 | c2gamA_ |
|
not modelled |
90.5 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:beta-1,6-n-acetylglucosaminyltransferase;
PDBTitle: x-ray crystal structure of murine leukocyte-type core 2 b1,6-n-2 acetylglucosaminyltransferase (c2gnt-l) in complex with galb1,3galnac
|
|
|
| 69 | c3f1cB_ |
|
not modelled |
90.4 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:putative 2-c-methyl-d-erythritol 4-phosphate
PDBTitle: crystal structure of 2-c-methyl-d-erythritol 4-phosphate2 cytidylyltransferase from listeria monocytogenes
|
|
|
| 70 | c3tqdA_ |
|
not modelled |
90.3 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:3-deoxy-manno-octulosonate cytidylyltransferase;
PDBTitle: structure of the 3-deoxy-d-manno-octulosonate cytidylyltransferase2 (kdsb) from coxiella burnetii
|
|
|
| 71 | c4mybA_ |
|
not modelled |
90.1 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: crystal structure of francisella tularensis 2-c-methyl-d-erythritol 4-2 phosphate cytidylyltransferase (ispd)
|
|
|
| 72 | d1i52a_ |
|
not modelled |
90.1 |
12 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 73 | d1vpaa_ |
|
not modelled |
89.3 |
7 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 74 | c3okrC_ |
|
not modelled |
89.3 |
17 |
PDB header:transferase
Chain: C: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;
PDBTitle: structure of mtb apo 2-c-methyl-d-erythritol 4-phosphate2 cytidyltransferase (ispd)
|
|
|
| 75 | d1e5ka_ |
|
not modelled |
89.3 |
14 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Molybdenum cofactor biosynthesis protein MobA |
|
|
| 76 | c3d98A_ |
|
not modelled |
89.0 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: crystal structure of glmu from mycobacterium tuberculosis, ligand-free2 form
|
|
|
| 77 | c2vshB_ |
|
not modelled |
88.8 |
6 |
PDB header:transferase
Chain: B: PDB Molecule:2-c-methyl-d-erythritol 4-phosphate
PDBTitle: synthesis of cdp-activated ribitol for teichoic acid2 precursors in streptococcus pneumoniae
|
|
|
| 78 | d1vgwa_ |
|
not modelled |
88.5 |
12 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 79 | d1h7ea_ |
|
not modelled |
88.5 |
12 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 80 | d1vica_ |
|
not modelled |
88.3 |
12 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 81 | d1iina_ |
|
not modelled |
88.2 |
8 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
|
|
| 82 | d1w55a1 |
|
not modelled |
86.3 |
12 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 83 | c1w57A_ |
|
not modelled |
86.2 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:ispd/ispf bifunctional enzyme;
PDBTitle: structure of the bifunctional ispdf from campylobacter2 jejuni containing zn
|
|
|
| 84 | c6bwhB_ |
|
not modelled |
85.6 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:2-phospho-l-lactate guanylyltransferase;
PDBTitle: crystal structure of mycoibacterium tuberculosis rv2983 in complex2 with pep
|
|
|
| 85 | c4jd0A_ |
|
not modelled |
84.5 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:nucleotidyl transferase;
PDBTitle: structure of the inositol-1-phosphate ctp transferase from t.2 maritima.
|
|
|
| 86 | c2xmhB_ |
|
not modelled |
83.0 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:ctp-inositol-1-phosphate cytidylyltransferase;
PDBTitle: the x-ray structure of ctp:inositol-1-phosphate cytidylyltransferase2 from archaeoglobus fulgidus
|
|
|
| 87 | d2oi6a2 |
|
not modelled |
81.0 |
9 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:UDP-glucose pyrophosphorylase |
|
|
| 88 | c3rsbB_ |
|
not modelled |
80.3 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:adenosylcobinamide-phosphate guanylyltransferase;
PDBTitle: structure of the archaeal gtp:adocbi-p guanylyltransferase (coby) from2 methanocaldococcus jannaschii
|
|
|
| 89 | c2qkxA_ |
|
not modelled |
79.0 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:bifunctional protein glmu;
PDBTitle: n-acetyl glucosamine 1-phosphate uridyltransferase from mycobacterium2 tuberculosis complex with n-acetyl glucosamine 1-phosphate
|
|
|
| 90 | c5xhwA_ |
|
not modelled |
78.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:putative 6-deoxy-d-mannoheptose pathway protein;
PDBTitle: crystal structure of hddc from yersinia pseudotuberculosis
|
|
|
| 91 | c4jisB_ |
|
not modelled |
78.7 |
6 |
PDB header:transferase
Chain: B: PDB Molecule:ribitol-5-phosphate cytidylyltransferase;
PDBTitle: crystal structure of ribitol 5-phosphate cytidylyltransferase (tari)2 from bacillus subtilis
|
|
|
| 92 | c2pa4B_ |
|
not modelled |
78.2 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:utp-glucose-1-phosphate uridylyltransferase;
PDBTitle: crystal structure of udp-glucose pyrophosphorylase from corynebacteria2 glutamicum in complex with magnesium and udp-glucose
|
|
|
| 93 | c6aokA_ |
|
not modelled |
76.7 |
31 |
PDB header:hydrolase
Chain: A: PDB Molecule:ceg4;
PDBTitle: crystal structure of legionella pneumophila effector ceg4 with n-2 terminal tev protease cleavage sequence
|
|
|
| 94 | c3polA_ |
|
not modelled |
76.3 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:3-deoxy-manno-octulosonate cytidylyltransferase;
PDBTitle: 2.3 angstrom crystal structure of 3-deoxy-manno-octulosonate2 cytidylyltransferase (kdsb) from acinetobacter baumannii.
|
|
|
| 95 | c6cgjA_ |
|
not modelled |
75.3 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:effector protein lem4 (lpg1101);
PDBTitle: structure of the had domain of effector protein lem4 (lpg1101) from2 legionella pneumophila
|
|
|
| 96 | d1eyra_ |
|
not modelled |
75.0 |
10 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 97 | d1vh1a_ |
|
not modelled |
67.4 |
11 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 98 | c2y6pC_ |
|
not modelled |
66.4 |
11 |
PDB header:transferase
Chain: C: PDB Molecule:3-deoxy-manno-octulosonate cytidylyltransferase;
PDBTitle: evidence for a two-metal-ion-mechanism in the kdo-2 cytidylyltransferase kdsb
|
|
|
| 99 | c3brkX_ |
|
not modelled |
64.4 |
12 |
PDB header:transferase
Chain: X: PDB Molecule:glucose-1-phosphate adenylyltransferase;
PDBTitle: crystal structure of adp-glucose pyrophosphorylase from agrobacterium2 tumefaciens
|
|
|
| 100 | c6ifdD_ |
|
not modelled |
64.3 |
7 |
PDB header:sugar binding protein
Chain: D: PDB Molecule:cmp-n-acetylneuraminate synthetase;
PDBTitle: crystal structure of cmp-n-acetylneuraminate synthetase from vibrio2 cholerae in complex with cdp and mg2+.
|
|
|
| 101 | c3brcA_ |
|
not modelled |
63.5 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved protein of unknown function;
PDBTitle: crystal structure of a conserved protein of unknown function from2 methanobacterium thermoautotrophicum
|
|
|
| 102 | c3pnnA_ |
|
not modelled |
63.4 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:conserved domain protein;
PDBTitle: the crystal structure of a glycosyltransferase from porphyromonas2 gingivalis w83
|
|
|
| 103 | c6qg2F_ |
|
not modelled |
61.9 |
11 |
PDB header:translation
Chain: F: PDB Molecule:translation initiation factor eif-2b subunit gamma;
PDBTitle: structure of eif2b-eif2 (phosphorylated at ser51) complex (model a)
|
|
|
| 104 | c3ngwA_ |
|
not modelled |
60.2 |
11 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:molybdopterin-guanine dinucleotide biosynthesis protein a
PDBTitle: crystal structure of molybdopterin-guanine dinucleotide biosynthesis2 protein a from archaeoglobus fulgidus, northeast structural genomics3 consortium target gr189
|
|
|
| 105 | c3ssmB_ |
|
not modelled |
60.0 |
27 |
PDB header:transferase
Chain: B: PDB Molecule:methyltransferase;
PDBTitle: myce methyltransferase from the mycinamycin biosynthetic pathway in2 complex with mg and sah, crystal form 1
|
|
|
| 106 | c6ejeA_ |
|
not modelled |
59.7 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:xylosyltransferase 1;
PDBTitle: human xylosyltransferase 1 in complex with peptide paaegsgeqdft
|
|
|
| 107 | c5d5pC_ |
|
not modelled |
59.6 |
18 |
PDB header:transferase
Chain: C: PDB Molecule:hcgb;
PDBTitle: hcgb from methanococcus maripaludis
|
|
|
| 108 | c3hl3A_ |
|
not modelled |
57.8 |
7 |
PDB header:transferase
Chain: A: PDB Molecule:glucose-1-phosphate thymidylyltransferase;
PDBTitle: 2.76 angstrom crystal structure of a putative glucose-1-phosphate2 thymidylyltransferase from bacillus anthracis in complex with a3 sucrose.
|
|
|
| 109 | c3ssoE_ |
|
not modelled |
57.1 |
26 |
PDB header:transferase
Chain: E: PDB Molecule:methyltransferase;
PDBTitle: myce methyltransferase from the mycinamycin biosynthetic pathway in2 complex with mg and sah, crystal form 2
|
|
|
| 110 | d2dpwa1 |
|
not modelled |
56.5 |
12 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:TTHA0179-like |
|
|
| 111 | c1jylC_ |
|
not modelled |
55.0 |
16 |
PDB header:transferase
Chain: C: PDB Molecule:ctp:phosphocholine cytidylytransferase;
PDBTitle: catalytic mechanism of ctp:phosphocholine2 cytidylytransferase from streptococcus pneumoniae (licc)
|
|
|
| 112 | d1g97a2 |
|
not modelled |
50.2 |
9 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:UDP-glucose pyrophosphorylase |
|
|
| 113 | c4mndA_ |
|
not modelled |
48.8 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:ctp l-myo-inositol-1-phosphate cytidylyltransferase/cdp-l-
PDBTitle: crystal structure of archaeoglobus fulgidus ipct-dipps bifunctional2 membrane protein
|
|
|
| 114 | c4uegB_ |
|
not modelled |
47.4 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:glycogenin-2;
PDBTitle: crystal structure of human glycogenin-2 catalytic domain
|
|
|
| 115 | c5yh1A_ |
|
not modelled |
47.1 |
5 |
PDB header:ribosomal protein
Chain: A: PDB Molecule:member of s1p family of ribosomal proteins;
PDBTitle: member of s1p family of ribosomal proteins pf0399 dhh domain
|
|
|
| 116 | c2x5sB_ |
|
not modelled |
45.8 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:mannose-1-phosphate guanylyltransferase;
PDBTitle: crystal structure of t. maritima gdp-mannose2 pyrophosphorylase in apo state.
|
|
|
| 117 | c4y7uA_ |
|
not modelled |
44.5 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:nucleotidyl transferase;
PDBTitle: structural analysis of muru
|
|
|
| 118 | c2ux8G_ |
|
not modelled |
44.4 |
18 |
PDB header:transferase
Chain: G: PDB Molecule:glucose-1-phosphate uridylyltransferase;
PDBTitle: crystal structure of sphingomonas elodea atcc 31461 glucose-2 1-phosphate uridylyltransferase in complex with glucose-3 1-phosphate.
|
|
|
| 119 | d1ll2a_ |
|
not modelled |
43.6 |
14 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Glycogenin |
|
|
| 120 | d1vkpa_ |
|
not modelled |
43.2 |
14 |
Fold:Pentein, beta/alpha-propeller
Superfamily:Pentein
Family:Porphyromonas-type peptidylarginine deiminase |
|
|