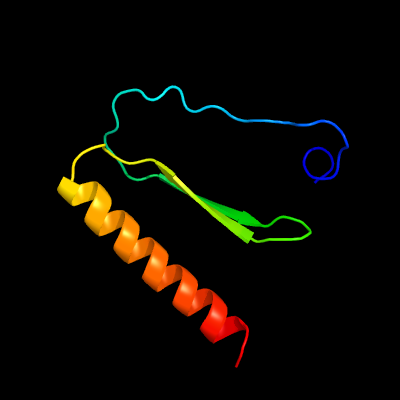
1 c6aefB_
98.3
36
PDB header: transferaseChain: B: PDB Molecule: polyketide synthase associated protein papa2;PDBTitle: papa2 acyl transferase
2 c6ehrF_
70.9
7
PDB header: signaling proteinChain: F: PDB Molecule: ras-related gtp-binding protein a;PDBTitle: the crystal structure of the human lamtor-raga ctd-ragc ctd complex
3 c2w91A_
36.6
38
PDB header: hydrolaseChain: A: PDB Molecule: endo-beta-n-acetylglucosaminidase d;PDBTitle: structure of a streptococcus pneumoniae family 85 glycoside2 hydrolase, endo-d.
4 c3gdbA_
34.9
38
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein spr0440;PDBTitle: crystal structure of spr0440 glycoside hydrolase domain,2 endo-d from streptococcus pneumoniae r6
5 d3e9va1
20.5
18
Fold: BTG domain-likeSuperfamily: BTG domain-likeFamily: BTG domain-like
6 c3fhaD_
17.9
60
PDB header: hydrolaseChain: D: PDB Molecule: endo-beta-n-acetylglucosaminidase;PDBTitle: structure of endo-beta-n-acetylglucosaminidase a
7 c5y88V_
16.9
23
PDB header: splicingChain: V: PDB Molecule: pre-mrna-splicing factor ntr2;PDBTitle: cryo-em structure of the intron-lariat spliceosome ready for2 disassembly from s.cerevisiae at 3.5 angstrom
8 c4gu4B_
16.3
17
PDB header: viral proteinChain: B: PDB Molecule: outer capsid protein sigma-1;PDBTitle: crystal structure of the t1l reovirus attachment protein sigma1 in2 complex with alpha-2,3-sialyllactose
9 c5dijA_
16.3
6
PDB header: unknown functionChain: A: PDB Molecule: tqaa;PDBTitle: the crystal structure of ct
10 c3rlcA_
13.8
23
PDB header: structural proteinChain: A: PDB Molecule: a1 protein;PDBTitle: crystal structure of the read-through domain from bacteriophage qbeta2 a1 protein, hexagonal crystal form
11 c5t3eA_
12.9
9
PDB header: ligaseChain: A: PDB Molecule: bacillamide synthetase heterocyclization domain;PDBTitle: crystal structure of a nonribosomal peptide synthetase2 heterocyclization domain.
12 d1kkea2
11.7
30
Fold: Virus attachment protein globular domainSuperfamily: Virus attachment protein globular domainFamily: Reovirus attachment protein sigma 1 head domain
13 d2z15a1
10.9
21
Fold: BTG domain-likeSuperfamily: BTG domain-likeFamily: BTG domain-like
14 c4chjA_
10.6
29
PDB header: cell cycleChain: A: PDB Molecule: imc sub-compartment protein isp3;PDBTitle: structure of inner membrane complex (imc) sub-compartment2 protein 3 (isp3) from toxoplasma gondii
15 d1r46a2
10.0
16
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
16 c4cu9A_
9.8
14
PDB header: hydrolaseChain: A: PDB Molecule: beta-galactosidase;PDBTitle: unravelling the multiple functions of the architecturally2 intricate streptococcus pneumoniae beta-galactosidase, bgaa
17 c2n8kA_
9.6
26
PDB header: toxinChain: A: PDB Molecule: u33-theraphotoxin-cg1b;PDBTitle: chemical shift assignments and structure determination for spider2 toxin, u33-theraphotoxin-cg1c
18 c3kwrA_
8.6
25
PDB header: rna binding proteinChain: A: PDB Molecule: putative rna-binding protein;PDBTitle: crystal structure of putative rna-binding protein (np_785364.1) from2 lactobacillus plantarum at 1.45 a resolution
19 d1a4ia1
8.6
25
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain
20 c5td6A_
8.5
9
PDB header: rna binding proteinChain: A: PDB Molecule: fog-3 protein;PDBTitle: c. elegans fog-3 btg/tob domain - h47n, c117a
21 c1kkeA_
not modelled
8.2
30
PDB header: viral proteinChain: A: PDB Molecule: sigma 1 protein;PDBTitle: crystal structure of reovirus attachment protein sigma12 trimer
22 c6av8A_
not modelled
7.8
80
PDB header: toxinChain: A: PDB Molecule: u5-theraphotoxin-hs1b 1;PDBTitle: exploring cystine dense peptide space to open a unique molecular2 toolbox
23 c2aapA_
not modelled
7.5
80
PDB header: toxinChain: A: PDB Molecule: jingzhaotoxin-vii;PDBTitle: solution structure of jingzhaotoxin-vii
24 c3qmlC_
not modelled
7.5
26
PDB header: chaperone/protein transportChain: C: PDB Molecule: nucleotide exchange factor sil1;PDBTitle: the structural analysis of sil1-bip complex reveals the mechanism for2 sil1 to function as a novel nucleotide exchange factor
25 c3ushB_
not modelled
7.2
35
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the q2s0r5 protein from salinibacter ruber,2 northeast structural genomics consortium target srr207
26 c4chmB_
not modelled
7.0
12
PDB header: cell cycleChain: B: PDB Molecule: imc sub-compartment protein isp1;PDBTitle: structure of inner membrane complex (imc) sub-compartment protein 12 (isp1) from toxoplasma gondii
27 c3l07B_
not modelled
6.4
21
PDB header: oxidoreductase,hydrolaseChain: B: PDB Molecule: bifunctional protein fold;PDBTitle: methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate2 cyclohydrolase, putative bifunctional protein fold from francisella3 tularensis.
28 c3s6xA_
not modelled
6.3
30
PDB header: viral proteinChain: A: PDB Molecule: outer capsid protein sigma-1;PDBTitle: structure of reovirus attachment protein sigma1 in complex with alpha-2 2,3-sialyllactose
29 c1a4iB_
not modelled
6.0
24
PDB header: oxidoreductaseChain: B: PDB Molecule: methylenetetrahydrofolate dehydrogenase /PDBTitle: human tetrahydrofolate dehydrogenase / cyclohydrolase
30 c5oltA_
not modelled
6.0
44
PDB header: transferaseChain: A: PDB Molecule: cellulose biosynthesis protein bcsg;PDBTitle: crystal structure of the extramembrane domain of the cellulose2 biosynthetic protein bcsg from salmonella typhimurium
31 d1a6qa1
not modelled
5.9
38
Fold: Another 3-helical bundleSuperfamily: Protein serine/threonine phosphatase 2C, C-terminal domainFamily: Protein serine/threonine phosphatase 2C, C-terminal domain
32 c2lq3A_
not modelled
5.4
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of syc0711_d from synechococcus sp., northeast2 structural genomics consortium (nesg) target snr212
33 c3e2sA_
not modelled
5.3
15
PDB header: oxidoreductaseChain: A: PDB Molecule: proline dehydrogenase;PDBTitle: crystal structure reduced puta86-630 mutant y540s complexed with l-2 proline
34 c5yvuA_
not modelled
5.3
38
PDB header: viral proteinChain: A: PDB Molecule: genome polyprotein;PDBTitle: crystal structures of unlinked full length ns3 from dengue virus2 provide insights into dynamics of protease domain
35 d1p42a1
not modelled
5.3
28
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase LpxC
36 c2n59A_
not modelled
5.3
67
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein csgh;PDBTitle: solution structure of r. palustris csgh
37 c4iohA_
not modelled
5.1
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: tll1086 protein;PDBTitle: crystal structure of the tll1086 protein from thermosynechococcus2 elongatus, northeast structural genomics consortium target ter258
38 c5i8iD_
not modelled
5.1
22
PDB header: hydrolaseChain: D: PDB Molecule: urea amidolyase;PDBTitle: crystal structure of the k. lactis urea amidolyase
39 d1o5wa2
not modelled
5.0
13
Fold: FAD-linked reductases, C-terminal domainSuperfamily: FAD-linked reductases, C-terminal domainFamily: L-aminoacid/polyamine oxidase
40 c2k1kA_
not modelled
5.0
100
PDB header: signaling proteinChain: A: PDB Molecule: ephrin type-a receptor 1;PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 4.3
41 c2k1kB_
not modelled
5.0
100
PDB header: signaling proteinChain: B: PDB Molecule: ephrin type-a receptor 1;PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 4.3
42 c2k1lA_
not modelled
5.0
100
PDB header: signaling proteinChain: A: PDB Molecule: ephrin type-a receptor 1;PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 6.3