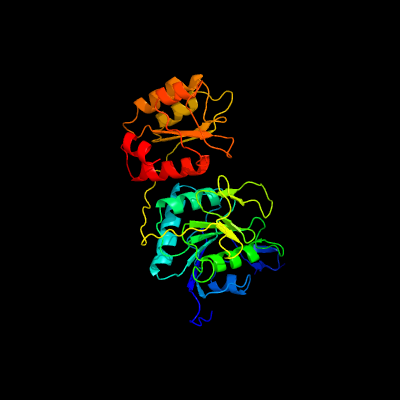
| 1 | c2d6fA_
|
|
 |
100.0 |
24 |
PDB header:ligase/rna
Chain: A: PDB Molecule:glutamyl-trna(gln) amidotransferase subunit d;
PDBTitle: crystal structure of glu-trna(gln) amidotransferase in the2 complex with trna(gln)
|
|
|
|
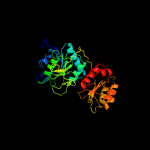
| 2 | c1zq1B_
|
|
 |
100.0 |
25 |
PDB header:lyase
Chain: B: PDB Molecule:glutamyl-trna(gln) amidotransferase subunit d;
PDBTitle: structure of gatde trna-dependent amidotransferase from2 pyrococcus abyssi
|
|
|
|
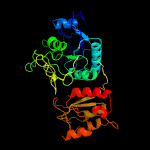
| 3 | d1zq1a2
|
|
 |
100.0 |
25 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
|
|
|
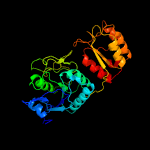
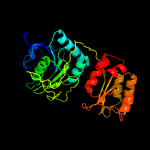
| 4 | d2d6fa2
|
|
 |
100.0 |
24 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
|
|
|
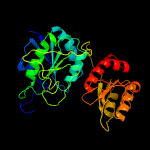
| 5 | d1nnsa_
|
|
 |
100.0 |
30 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
|
|
|
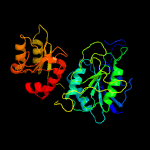
| 6 | d1agxa_
|
|
 |
100.0 |
26 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
|
|
|
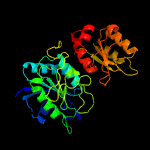
| 7 | c2wltA_
|
|
 |
100.0 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:l-asparaginase;
PDBTitle: the crystal structure of helicobacter pylori l-asparaginase at 1.4 a2 resolution
|
|
|
|
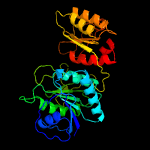
| 8 | d1wsaa_
|
|
 |
100.0 |
30 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
|
|
|
| 9 | d4pgaa_
|
|
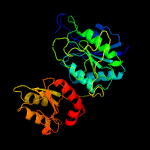 |
100.0 |
28 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
|
|
|
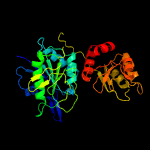
| 10 | c4q0mA_
|
|
 |
100.0 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:l-asparaginase;
PDBTitle: crystal structure of pyrococcus furiosus l-asparaginase
|
|
|
|
| 11 | d1o7ja_
|
|
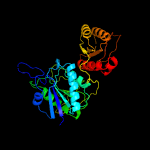 |
100.0 |
30 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
|
|
|
| 12 | c4r8kC_
|
|
 |
100.0 |
26 |
PDB header:hydrolase
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the guinea pig l-asparaginase 1 catalytic domain
|
|
|
|
| 13 | c1wnfA_
|
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:l-asparaginase;
PDBTitle: crystal structure of ph0066 from pyrococcus horikoshii
|
|
|
|
| 14 | d2ocda1
|
|
 |
100.0 |
24 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
|
|
|
| 15 | c3nxkE_
|
|
 |
100.0 |
30 |
PDB header:hydrolase
Chain: E: PDB Molecule:cytoplasmic l-asparaginase;
PDBTitle: crystal structure of probable cytoplasmic l-asparaginase from2 campylobacter jejuni
|
|
|
|
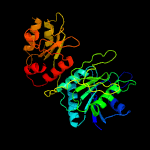
| 16 | c2p2dA_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:l-asparaginase i;
PDBTitle: crystal structure and allosteric regulation of the cytoplasmic2 escherichia coli l-asparaginase i
|
|
|
|
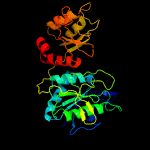
| 17 | c5ot0A_
|
|
 |
100.0 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:l-asparaginase;
PDBTitle: the thermostable l-asparaginase from thermococcus kodakarensis
|
|
|
|
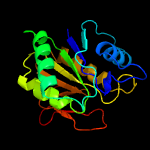
| 18 | c4ra6A_
|
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:l-asparaginase;
PDBTitle: crystal structure of linker less pyrococcus furiosus l-asparaginase
|
|
|
|
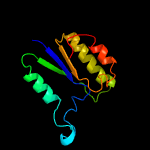
| 19 | c6cv6L_
|
|
 |
94.5 |
21 |
PDB header:lyase
Chain: L: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of 3-dehydroquinate dehydratase, type ii, from2 burkholderia phymatum stm815
|
|
|
|
| 20 | c3kipU_
|
|
 |
93.7 |
22 |
PDB header:lyase
Chain: U: PDB Molecule:3-dehydroquinase, type ii;
PDBTitle: crystal structure of type-ii 3-dehydroquinase from c. albicans
|
|
|
|
| 21 | d1uqra_ |
|
not modelled |
92.0 |
15 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
|
|
| 22 | c3s81A_ |
|
not modelled |
90.6 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:putative aspartate racemase;
PDBTitle: crystal structure of putative aspartate racemase from salmonella2 typhimurium
|
|
|
| 23 | d1gqoa_ |
|
not modelled |
87.8 |
14 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
|
|
| 24 | c2ad5B_ |
|
not modelled |
85.6 |
17 |
PDB header:ligase
Chain: B: PDB Molecule:ctp synthase;
PDBTitle: mechanisms of feedback regulation and drug resistance of ctp2 synthetases: structure of the e. coli ctps/ctp complex at 2.8-3 angstrom resolution.
|
|
|
| 25 | c5lg5F_ |
|
not modelled |
85.1 |
20 |
PDB header:isomerase
Chain: F: PDB Molecule:allantoin racemase;
PDBTitle: crystal structure of allantoin racemase from pseudomonas fluorescens2 allr
|
|
|
| 26 | c3qvjB_ |
|
not modelled |
84.2 |
24 |
PDB header:isomerase
Chain: B: PDB Molecule:putative hydantoin racemase;
PDBTitle: allantoin racemase from klebsiella pneumoniae
|
|
|
| 27 | c4yajA_ |
|
not modelled |
81.9 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:alpha subunit of acetyl-coenzyme a synthetase
PDBTitle: ca. korarchaeum cryptofilum dinucleotide forming acetyl-coenzyme a2 synthetase 1 (apo form)
|
|
|
| 28 | c5u03C_ |
|
not modelled |
80.1 |
20 |
PDB header:ligase, protein fibril
Chain: C: PDB Molecule:ctp synthase 1;
PDBTitle: cryo-em structure of the human ctp synthase filament
|
|
|
| 29 | c4zdiE_ |
|
not modelled |
78.7 |
19 |
PDB header:ligase
Chain: E: PDB Molecule:ctp synthase;
PDBTitle: crystal structure of the m. tuberculosis ctp synthase pyrg (apo form)
|
|
|
| 30 | c3nvaB_ |
|
not modelled |
77.9 |
14 |
PDB header:ligase
Chain: B: PDB Molecule:ctp synthase;
PDBTitle: dimeric form of ctp synthase from sulfolobus solfataricus
|
|
|
| 31 | c3oa0B_ |
|
not modelled |
77.8 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:lipopolysaccaride biosynthesis protein wbpb;
PDBTitle: crystal structure of the wlba (wbpb) dehydrogenase from thermus2 thermophilus in complex with nad and udp-glcnaca
|
|
|
| 32 | c4p5pA_ |
|
not modelled |
77.5 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:thij/pfpi family protein;
PDBTitle: x-ray structure of francisella tularensis rapid encystment protein 242 kda (rep24), gene product of ftn_0841
|
|
|
| 33 | c1uz5A_ |
|
not modelled |
76.3 |
18 |
PDB header:molybdopterin biosynthesis
Chain: A: PDB Molecule:402aa long hypothetical molybdopterin
PDBTitle: the crystal structure of molybdopterin biosynthesis moea2 protein from pyrococcus horikosii
|
|
|
| 34 | d1pfka_ |
|
not modelled |
75.4 |
20 |
Fold:Phosphofructokinase
Superfamily:Phosphofructokinase
Family:Phosphofructokinase |
|
|
| 35 | d1uz5a3 |
|
not modelled |
74.2 |
19 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
|
|
| 36 | c2nqqA_ |
|
not modelled |
74.2 |
22 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:molybdopterin biosynthesis protein moea;
PDBTitle: moea r137q
|
|
|
| 37 | c2zskA_ |
|
not modelled |
73.6 |
19 |
PDB header:unknown function
Chain: A: PDB Molecule:226aa long hypothetical aspartate racemase;
PDBTitle: crystal structure of ph1733, an aspartate racemase2 homologue, from pyrococcus horikoshii ot3
|
|
|
| 38 | d2ftsa3 |
|
not modelled |
69.7 |
13 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
|
|
| 39 | d2c4va1 |
|
not modelled |
69.6 |
15 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
|
|
| 40 | c4rhcH_ |
|
not modelled |
68.7 |
16 |
PDB header:lyase
Chain: H: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of 3-dehydroquinate dehydratase from acinetobacter2 baumannii at 2.68 a resolution
|
|
|
| 41 | c2h2wA_ |
|
not modelled |
68.5 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:homoserine o-succinyltransferase;
PDBTitle: crystal structure of homoserine o-succinyltransferase (ec 2.3.1.46)2 (homoserine o-transsuccinylase) (hts) (tm0881) from thermotoga3 maritima at 2.52 a resolution
|
|
|
| 42 | c2dx7B_ |
|
not modelled |
66.4 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:aspartate racemase;
PDBTitle: crystal structure of pyrococcus horikoshii ot3 aspartate racemase2 complex with citric acid
|
|
|
| 43 | c4wpgA_ |
|
not modelled |
66.1 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dtdp-4-dehydrorhamnose reductase;
PDBTitle: group a streptococcus gaca is an essential dtdp-4-dehydrorhamnose2 reductase (rmld)
|
|
|
| 44 | d2nqra3 |
|
not modelled |
63.9 |
23 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
|
|
| 45 | c5l3rC_ |
|
not modelled |
63.1 |
17 |
PDB header:protein transport
Chain: C: PDB Molecule:signal recognition particle 54 kda protein, chloroplastic;
PDBTitle: structure of the gtpase heterodimer of chloroplast srp54 and ftsy from2 arabidopsis thaliana
|
|
|
| 46 | c5elmB_ |
|
not modelled |
60.3 |
19 |
PDB header:isomerase
Chain: B: PDB Molecule:asp/glu_racemase family protein;
PDBTitle: crystal structure of l-aspartate/glutamate specific racemase in2 complex with l-glutamate
|
|
|
| 47 | c5xzdF_ |
|
not modelled |
60.2 |
11 |
PDB header:lyase
Chain: F: PDB Molecule:enoyl-coa hydratase;
PDBTitle: structure of acryloyl-coa hydratase acuh from roseovarius nubinhibens2 ism
|
|
|
| 48 | c2uygF_ |
|
not modelled |
60.1 |
18 |
PDB header:lyase
Chain: F: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystallogaphic structure of the typeii 3-dehydroquinase from thermus2 thermophilus
|
|
|
| 49 | c6mtgB_ |
|
not modelled |
59.4 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:homoserine o-succinyltransferase;
PDBTitle: a single reactive noncanonical amino acid is able to dramatically2 stabilize protein structure
|
|
|
| 50 | c5wxzA_ |
|
not modelled |
57.8 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:mcyf;
PDBTitle: crystal structure of microcystis aeruginosa pcc 7806 aspartate2 racemase in complex with d-aspartate
|
|
|
| 51 | d1ls1a2 |
|
not modelled |
57.3 |
19 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
|
|
| 52 | c3e96B_ |
|
not modelled |
56.9 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from bacillus2 clausii
|
|
|
| 53 | c5g2rA_ |
|
not modelled |
56.5 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:molybdopterin biosynthesis protein cnx1;
PDBTitle: crystal structure of the mo-insertase domain cnx1e from2 arabidopsis thaliana
|
|
|
| 54 | d1tuba1 |
|
not modelled |
53.3 |
13 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
|
|
| 55 | c3lwzC_ |
|
not modelled |
53.2 |
15 |
PDB header:lyase
Chain: C: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: 1.65 angstrom resolution crystal structure of type ii 3-dehydroquinate2 dehydratase (aroq) from yersinia pestis
|
|
|
| 56 | c3s29C_ |
|
not modelled |
52.8 |
13 |
PDB header:transferase
Chain: C: PDB Molecule:sucrose synthase 1;
PDBTitle: the crystal structure of sucrose synthase-1 from arabidopsis thaliana2 and its functional implications.
|
|
|
| 57 | d2csua3 |
|
not modelled |
51.4 |
12 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
|
|
| 58 | c1powA_ |
|
not modelled |
51.0 |
13 |
PDB header:oxidoreductase(oxygen as acceptor)
Chain: A: PDB Molecule:pyruvate oxidase;
PDBTitle: the refined structures of a stabilized mutant and of wild-type2 pyruvate oxidase from lactobacillus plantarum
|
|
|
| 59 | c4l8lA_ |
|
not modelled |
49.7 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate dehydratase 1;
PDBTitle: crystal structure of the type ii dehydroquinase from pseudomonas2 aeruginosa
|
|
|
| 60 | c3oc7A_ |
|
not modelled |
49.3 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:enoyl-coa hydratase;
PDBTitle: crystal structure of an enoyl-coa hydratase from mycobacterium avium
|
|
|
| 61 | d1uuya_ |
|
not modelled |
48.6 |
23 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
|
|
| 62 | c2dwuA_ |
|
not modelled |
47.2 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase isoform race1 from bacillus2 anthracis
|
|
|
| 63 | c4lk5B_ |
|
not modelled |
46.6 |
25 |
PDB header:lyase
Chain: B: PDB Molecule:enoyl-coa hydratase;
PDBTitle: crystal structure of a enoyl-coa hydratase from mycobacterium avium2 subsp. paratuberculosis k-10
|
|
|
| 64 | c2jfoB_ |
|
not modelled |
46.4 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of enterococcus faecalis glutamate2 racemase in complex with d- and l-glutamate
|
|
|
| 65 | c3u7rB_ |
|
not modelled |
45.3 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadph-dependent fmn reductase;
PDBTitle: ferb - flavoenzyme nad(p)h:(acceptor) oxidoreductase (ferb) from2 paracoccus denitrificans
|
|
|
| 66 | d2f48a1 |
|
not modelled |
45.0 |
15 |
Fold:Phosphofructokinase
Superfamily:Phosphofructokinase
Family:Phosphofructokinase |
|
|
| 67 | d1wu2a3 |
|
not modelled |
44.7 |
21 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
|
|
| 68 | c5kt0A_ |
|
not modelled |
44.3 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-tetrahydrodipicolinate reductase;
PDBTitle: dihydrodipicolinate reductase from the industrial and evolutionarily2 important cyanobacteria anabaena variabilis.
|
|
|
| 69 | d1s2da_ |
|
not modelled |
44.1 |
18 |
Fold:Flavodoxin-like
Superfamily:N-(deoxy)ribosyltransferase-like
Family:N-deoxyribosyltransferase |
|
|
| 70 | c2eq5D_ |
|
not modelled |
43.6 |
26 |
PDB header:isomerase
Chain: D: PDB Molecule:228aa long hypothetical hydantoin racemase;
PDBTitle: crystal structure of hydantoin racemase from pyrococcus horikoshii ot3
|
|
|
| 71 | d1jlja_ |
|
not modelled |
42.6 |
17 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
|
|
| 72 | d1b74a1 |
|
not modelled |
41.9 |
13 |
Fold:ATC-like
Superfamily:Aspartate/glutamate racemase
Family:Aspartate/glutamate racemase |
|
|
| 73 | d1u9ca_ |
|
not modelled |
41.7 |
21 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:DJ-1/PfpI |
|
|
| 74 | c3h02F_ |
|
not modelled |
41.4 |
19 |
PDB header:lyase
Chain: F: PDB Molecule:naphthoate synthase;
PDBTitle: 2.15 angstrom resolution crystal structure of naphthoate synthase from2 salmonella typhimurium.
|
|
|
| 75 | c3c5yD_ |
|
not modelled |
41.3 |
16 |
PDB header:isomerase
Chain: D: PDB Molecule:ribose/galactose isomerase;
PDBTitle: crystal structure of a putative ribose 5-phosphate isomerase2 (saro_3514) from novosphingobium aromaticivorans dsm at 1.81 a3 resolution
|
|
|
| 76 | c3mc3A_ |
|
not modelled |
40.6 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dsre/dsrf-like family protein;
PDBTitle: crystal structure of dsre/dsrf-like family protein (np_342589.1) from2 sulfolobus solfataricus at 1.49 a resolution
|
|
|
| 77 | c4wzzA_ |
|
not modelled |
40.0 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:putative sugar abc transporter, substrate-binding protein;
PDBTitle: crystal structure of an abc transporter solute binding protein2 (ipr025997) from clostridium phytofermentas (cphy_0583, target efi-3 511148) with bound l-rhamnose
|
|
|
| 78 | c3b9qA_ |
|
not modelled |
38.3 |
13 |
PDB header:protein transport
Chain: A: PDB Molecule:chloroplast srp receptor homolog, alpha subunit
PDBTitle: the crystal structure of cpftsy from arabidopsis thaliana
|
|
|
| 79 | c5jp6A_ |
|
not modelled |
37.8 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative polysaccharide deacetylase;
PDBTitle: bdellovibrio bacteriovorus peptidoglycan deacetylase bd3279
|
|
|
| 80 | d1rjma_ |
|
not modelled |
37.3 |
15 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Crotonase-like |
|
|
| 81 | c3hfrA_ |
|
not modelled |
36.9 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase from listeria monocytogenes
|
|
|
| 82 | c2qs0A_ |
|
not modelled |
36.5 |
17 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:quinolinate synthetase a;
PDBTitle: quinolinate synthase from pyrococcus furiosus
|
|
|
| 83 | d1fuia2 |
|
not modelled |
36.0 |
13 |
Fold:FucI/AraA N-terminal and middle domains
Superfamily:FucI/AraA N-terminal and middle domains
Family:L-fucose isomerase, N-terminal and second domains |
|
|
| 84 | c2hmcA_ |
|
not modelled |
35.8 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: the crystal structure of dihydrodipicolinate synthase dapa from2 agrobacterium tumefaciens
|
|
|
| 85 | d1xi8a3 |
|
not modelled |
35.5 |
23 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
|
|
| 86 | d1mkza_ |
|
not modelled |
35.2 |
19 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
|
|
| 87 | c4m1bA_ |
|
not modelled |
34.7 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:polysaccharide deacetylase;
PDBTitle: structural determination of ba0150, a polysaccharide deacetylase from2 bacillus anthracis
|
|
|
| 88 | c3uhfB_ |
|
not modelled |
34.2 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase from campylobacter jejuni2 subsp. jejuni
|
|
|
| 89 | c2is8A_ |
|
not modelled |
33.9 |
20 |
PDB header:structural protein
Chain: A: PDB Molecule:molybdopterin biosynthesis enzyme, moab;
PDBTitle: crystal structure of the molybdopterin biosynthesis enzyme moab2 (ttha0341) from thermus theromophilus hb8
|
|
|
| 90 | d1c2ya_ |
|
not modelled |
33.6 |
17 |
Fold:Lumazine synthase
Superfamily:Lumazine synthase
Family:Lumazine synthase |
|
|
| 91 | d2g2ca1 |
|
not modelled |
33.5 |
23 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
|
|
| 92 | c4ak9A_ |
|
not modelled |
33.3 |
12 |
PDB header:protein transport
Chain: A: PDB Molecule:cpftsy;
PDBTitle: structure of chloroplast ftsy from physcomitrella patens
|
|
|
| 93 | c3bq9A_ |
|
not modelled |
32.8 |
44 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted rossmann fold nucleotide-binding domain-
PDBTitle: crystal structure of predicted nucleotide-binding protein from2 idiomarina baltica os145
|
|
|
| 94 | c3qreA_ |
|
not modelled |
32.7 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:enoyl-coa hydratase, echa12_1;
PDBTitle: crystal structure of an enoyl-coa hydratase echa12_1 from2 mycobacterium marinum
|
|
|
| 95 | c2jfqA_ |
|
not modelled |
32.5 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of staphylococcus aureus glutamate racemase in2 complex with d- glutamate
|
|
|
| 96 | c3p85A_ |
|
not modelled |
32.5 |
25 |
PDB header:lyase
Chain: A: PDB Molecule:enoyl-coa hydratase;
PDBTitle: crystal structure enoyl-coa hydratase from mycobacterium avium
|
|
|
| 97 | c2jfnA_ |
|
not modelled |
32.2 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of escherichia coli glutamate racemase2 in complex with l-glutamate and activator udp-murnac-ala
|
|
|
| 98 | d2nu7b1 |
|
not modelled |
31.7 |
17 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
|
|
| 99 | d1nqua_ |
|
not modelled |
31.5 |
24 |
Fold:Lumazine synthase
Superfamily:Lumazine synthase
Family:Lumazine synthase |
|
|
| 100 | d2f7wa1 |
|
not modelled |
31.5 |
19 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
|
|
| 101 | c1zxxA_ |
|
not modelled |
31.3 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:6-phosphofructokinase;
PDBTitle: the crystal structure of phosphofructokinase from lactobacillus2 delbrueckii
|
|
|
| 102 | d1vmaa2 |
|
not modelled |
31.3 |
11 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
|
|
| 103 | c2jfzB_ |
|
not modelled |
31.2 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of helicobacter pylori glutamate racemase2 in complex with d-glutamate and an inhibitor
|
|
|
| 104 | c4mi2C_ |
|
not modelled |
31.2 |
22 |
PDB header:isomerase
Chain: C: PDB Molecule:putative enoyl-coa hydratase/isomerase;
PDBTitle: crystal structure of putative enoyl-coa hydratase/isomerase from2 mycobacterium abscessus
|
|
|
| 105 | c5xvsA_ |
|
not modelled |
30.5 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:gdp/udp-n,n'-diacetylbacillosamine 2-epimerase
PDBTitle: crystal structure of udp-glcnac 2-epimerase neuc complexed with udp
|
|
|
| 106 | d1wzua1 |
|
not modelled |
30.4 |
25 |
Fold:NadA-like
Superfamily:NadA-like
Family:NadA-like |
|
|
| 107 | c2og2A_ |
|
not modelled |
29.3 |
13 |
PDB header:protein transport
Chain: A: PDB Molecule:putative signal recognition particle receptor;
PDBTitle: crystal structure of chloroplast ftsy from arabidopsis2 thaliana
|
|
|
| 108 | c2qjhH_ |
|
not modelled |
29.3 |
15 |
PDB header:lyase
Chain: H: PDB Molecule:putative aldolase mj0400;
PDBTitle: m. jannaschii adh synthase covalently bound to2 dihydroxyacetone phosphate
|
|
|
| 109 | c4jenB_ |
|
not modelled |
29.2 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:cmp n-glycosidase;
PDBTitle: structure of clostridium botulinum cmp n-glycosidase, bcmb
|
|
|
| 110 | c2iexA_ |
|
not modelled |
28.9 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:dihydroxynapthoic acid synthetase;
PDBTitle: crystal structure of dihydroxynapthoic acid synthetase (gk2873) from2 geobacillus kaustophilus hta426
|
|
|
| 111 | c2gzmB_ |
|
not modelled |
28.6 |
20 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of the glutamate racemase from bacillus anthracis
|
|
|
| 112 | c2j7pA_ |
|
not modelled |
28.3 |
14 |
PDB header:signal recognition
Chain: A: PDB Molecule:signal recognition particle protein;
PDBTitle: gmppnp-stabilized ng domain complex of the srp gtpases ffh2 and ftsy
|
|
|
| 113 | c6dspB_ |
|
not modelled |
28.3 |
16 |
PDB header:signaling protein
Chain: B: PDB Molecule:autoinducer 2-binding protein lsrb;
PDBTitle: lsrb from clostridium saccharobutylicum in complex with ai-2
|
|
|
| 114 | c4r1oF_ |
|
not modelled |
28.3 |
16 |
PDB header:isomerase
Chain: F: PDB Molecule:l-arabinose isomerase;
PDBTitle: crystal structure of thermophilic geobacillus kaustophilus l-arabinose2 isomerase
|
|
|
| 115 | d1h05a_ |
|
not modelled |
28.2 |
24 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
|
|
| 116 | d1fs2b1 |
|
not modelled |
28.0 |
5 |
Fold:Skp1 dimerisation domain-like
Superfamily:Skp1 dimerisation domain-like
Family:Skp1 dimerisation domain-like |
|
|
| 117 | c5w16D_ |
|
not modelled |
27.8 |
16 |
PDB header:isomerase
Chain: D: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase from thermus thermophilus in2 complex with d-glutamate
|
|
|
| 118 | c4zu2A_ |
|
not modelled |
27.8 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative isohexenylglutaconyl-coa hydratase;
PDBTitle: pseudomonas aeruginosa atue
|
|
|
| 119 | c3fduF_ |
|
not modelled |
27.5 |
22 |
PDB header:isomerase
Chain: F: PDB Molecule:putative enoyl-coa hydratase/isomerase;
PDBTitle: crystal structure of a putative enoyl-coa hydratase/isomerase from2 acinetobacter baumannii
|
|
|
| 120 | c2ohoA_ |
|
not modelled |
27.2 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: structural basis for glutamate racemase inhibitor
|
|
|