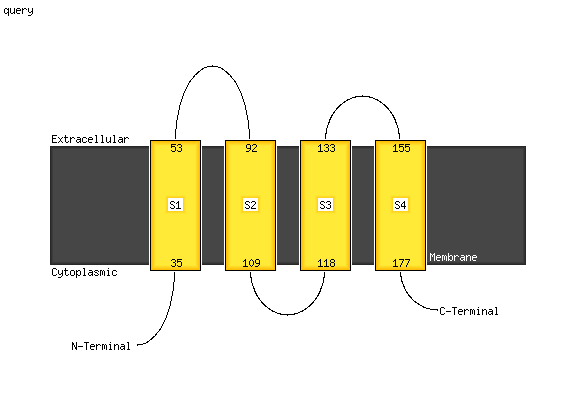
1 c5dirD_
100.0
38
PDB header: hydrolaseChain: D: PDB Molecule: lipoprotein signal peptidase;PDBTitle: membrane protein at 2.8 angstroms
2 c5dirB_
100.0
36
PDB header: hydrolaseChain: B: PDB Molecule: lipoprotein signal peptidase;PDBTitle: membrane protein at 2.8 angstroms
3 c6btmC_
83.7
11
PDB header: membrane proteinChain: C: PDB Molecule: alternative complex iii subunit c;PDBTitle: structure of alternative complex iii from flavobacterium johnsoniae2 (wild type)
4 c2kncA_
43.1
25
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
5 d2oara1
36.5
23
Fold: Gated mechanosensitive channelSuperfamily: Gated mechanosensitive channelFamily: Gated mechanosensitive channel
6 c3qnqD_
26.3
20
PDB header: membrane protein, transport proteinChain: D: PDB Molecule: pts system, cellobiose-specific iic component;PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
7 c2mfrA_
18.7
23
PDB header: transferaseChain: A: PDB Molecule: insulin receptor;PDBTitle: solution structure of the transmembrane domain of the insulin receptor2 in micelles
8 c6f0kC_
16.8
13
PDB header: membrane proteinChain: C: PDB Molecule: polysulphide reductase nrfd;PDBTitle: alternative complex iii
9 c5ireA_
16.7
24
PDB header: virusChain: A: PDB Molecule: e protein;PDBTitle: the cryo-em structure of zika virus
10 c6fkip_
15.8
7
PDB header: membrane proteinChain: P: PDB Molecule: atp synthase subunit c, chloroplastic;PDBTitle: chloroplast f1fo conformation 3
11 c5v8kB_
14.0
56
PDB header: photosynthesisChain: B: PDB Molecule: proteinsubunit pshx;PDBTitle: homodimeric reaction center of h. modesticaldum
12 c6iuhC_
12.0
33
PDB header: protein bindingChain: C: PDB Molecule: liprin-alpha-2;PDBTitle: crystal structure of git1 pbd domain in complex with liprin-alpha2
13 c3rb8A_
11.9
19
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: structure of the phage tubulin phuz(semet)-gdp
14 c6btmF_
10.6
9
PDB header: membrane proteinChain: F: PDB Molecule: alternative complex iii subunit f;PDBTitle: structure of alternative complex iii from flavobacterium johnsoniae2 (wild type)
15 d2d5ba1
10.2
46
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases
16 c2d46A_
10.1
37
PDB header: metal transportChain: A: PDB Molecule: calcium channel, voltage-dependent, beta 4PDBTitle: solution structure of the human beta4a-a domain
17 c2oarA_
9.6
23
PDB header: membrane proteinChain: A: PDB Molecule: large-conductance mechanosensitive channel;PDBTitle: mechanosensitive channel of large conductance (mscl)
18 c6f0kF_
9.1
16
PDB header: membrane proteinChain: F: PDB Molecule: actf;PDBTitle: alternative complex iii
19 c6oitG_
8.4
26
PDB header: plant proteinChain: G: PDB Molecule: protein chromatin remodeling 35;PDBTitle: cryoem structure of arabidopsis ddr' complex (drd1 peptide-dms3-rdm1)
20 c3hzqA_
7.8
15
PDB header: membrane proteinChain: A: PDB Molecule: large-conductance mechanosensitive channel;PDBTitle: structure of a tetrameric mscl in an expanded intermediate state
21 c3uzeC_
not modelled
7.4
21
PDB header: immune systemChain: C: PDB Molecule: envelope protein;PDBTitle: crystal structure of the dengue virus serotype 3 envelope protein2 domain iii in complex with the variable domains of mab 4e11
22 c4y7jE_
not modelled
7.3
32
PDB header: membrane protein,transport proteinChain: E: PDB Molecule: large conductance mechanosensitive channel protein,PDBTitle: structure of an archaeal mechanosensitive channel in expanded state
23 d1s7ba_
not modelled
7.3
9
Fold: Multidrug resistance efflux transporter EmrESuperfamily: Multidrug resistance efflux transporter EmrEFamily: Multidrug resistance efflux transporter EmrE
24 c3b9bA_
not modelled
6.9
23
PDB header: hydrolaseChain: A: PDB Molecule: sarcoplasmic/endoplasmic reticulum calciumPDBTitle: structure of the e2 beryllium fluoride complex of the serca2 ca2+-atpase
25 c1p58C_
not modelled
6.7
16
PDB header: virusChain: C: PDB Molecule: major envelope protein e;PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
26 c6iz4G_
not modelled
6.3
20
PDB header: membrane proteinChain: G: PDB Molecule: trimeric intracellular cation channel type b-b;PDBTitle: crystal structure analysis of tric counter-ion channels in calcium2 release
27 c2voyC_
not modelled
5.8
28
PDB header: hydrolaseChain: C: PDB Molecule: sarcoplasmic/endoplasmic reticulum calcium atpase 1;PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
28 d1ft8e_
not modelled
5.8
11
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Non-canonical RBD domain
29 c1x4pA_
not modelled
5.6
28
PDB header: rna binding proteinChain: A: PDB Molecule: putative splicing factor, arginine/serine-richPDBTitle: solution structure of surp domain in sfrs14 protei