| 1 |
|
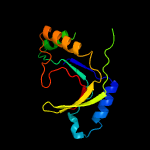
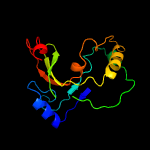
PDB 2d4g chain A
Region: 1 - 166
Aligned: 153
Modelled: 151
Confidence: 99.9%
Identity: 16%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein bsu11850;
PDBTitle: structure of yjcg protein, a putative 2'-5' rna ligase from bacillus2 subtilis
Phyre2
| 2 |
|
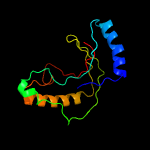
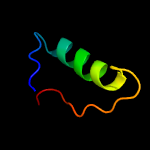
PDB 1jh6 chain A
Region: 2 - 167
Aligned: 154
Modelled: 154
Confidence: 99.9%
Identity: 16%
Fold: LigT-like
Superfamily: LigT-like
Family: tRNA splicing product Appr>p cyclic nucleotide phosphodiesterase
Phyre2
| 3 |
|
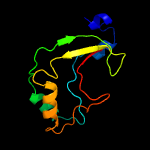
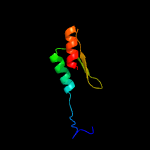
PDB 4qak chain A
Region: 5 - 167
Aligned: 151
Modelled: 163
Confidence: 99.9%
Identity: 14%
PDB header:hydrolase
Chain: A: PDB Molecule:2'-5'-rna ligase;
PDBTitle: crystal structure of phosphoesterase
Phyre2
| 4 |
|
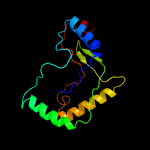
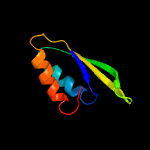
PDB 1vdx chain A
Region: 1 - 167
Aligned: 161
Modelled: 167
Confidence: 99.9%
Identity: 12%
PDB header:ligase
Chain: A: PDB Molecule:hypothetical protein ph0099;
PDBTitle: crystal structure of a pyrococcus horikoshii protein with2 similarities to 2'5' rna-ligase
Phyre2
| 5 |
|
PDB 5h7e chain A
Region: 5 - 167
Aligned: 151
Modelled: 163
Confidence: 99.9%
Identity: 19%
PDB header:hydrolase
Chain: A: PDB Molecule:rna 2',3'-cyclic phosphodiesterase;
PDBTitle: crystal structure of native drcpdase
Phyre2
| 6 |
|
PDB 1iuh chain A
Region: 1 - 167
Aligned: 162
Modelled: 162
Confidence: 99.9%
Identity: 14%
Fold: LigT-like
Superfamily: LigT-like
Family: 2'-5' RNA ligase LigT
Phyre2
| 7 |
|
PDB 2vfy chain A
Region: 1 - 128
Aligned: 122
Modelled: 128
Confidence: 99.8%
Identity: 13%
PDB header:hydrolase
Chain: A: PDB Molecule:akap18 delta;
PDBTitle: akap18 delta central domain
Phyre2
| 8 |
|
PDB 3j4r chain A
Region: 3 - 128
Aligned: 120
Modelled: 126
Confidence: 99.8%
Identity: 13%
PDB header:transferase
Chain: A: PDB Molecule:a-kinase anchor protein 18;
PDBTitle: pseudo-atomic model of the akap18-pka complex in a linear conformation2 derived from electron microscopy
Phyre2
| 9 |
|
PDB 4h7w chain A
Region: 5 - 128
Aligned: 118
Modelled: 124
Confidence: 99.7%
Identity: 14%
PDB header:unknown function
Chain: A: PDB Molecule:upf0406 protein c16orf57;
PDBTitle: crystal structure of human c16orf57
Phyre2
| 10 |
|
PDB 4z5v chain A
Region: 19 - 128
Aligned: 99
Modelled: 110
Confidence: 97.9%
Identity: 20%
PDB header:viral protein
Chain: A: PDB Molecule:non-structural protein 2a;
PDBTitle: crystal structure of mhv ns2 pde domain
Phyre2
| 11 |
|
PDB 2fsq chain A domain 1
Region: 4 - 143
Aligned: 135
Modelled: 138
Confidence: 93.7%
Identity: 18%
Fold: LigT-like
Superfamily: LigT-like
Family: Atu0111-like
Phyre2
| 12 |
|
PDB 5uqj chain A
Region: 5 - 142
Aligned: 133
Modelled: 138
Confidence: 87.5%
Identity: 15%
PDB header:hydrolase
Chain: A: PDB Molecule:u6 snrna phosphodiesterase;
PDBTitle: structure of yeast usb1
Phyre2
| 13 |
|
PDB 1em8 chain A
Region: 82 - 127
Aligned: 33
Modelled: 33
Confidence: 48.9%
Identity: 15%
Fold: DNA polymerase III chi subunit
Superfamily: DNA polymerase III chi subunit
Family: DNA polymerase III chi subunit
Phyre2
| 14 |
|
PDB 3gr1 chain A
Region: 1 - 61
Aligned: 61
Modelled: 61
Confidence: 35.3%
Identity: 10%
PDB header:membrane protein
Chain: A: PDB Molecule:protein prgh;
PDBTitle: periplasmic domain of the t3ss inner membrane protein prgh from2 s.typhimurium (fragment 170-392)
Phyre2
| 15 |
|
PDB 3ce8 chain A
Region: 40 - 108
Aligned: 69
Modelled: 69
Confidence: 34.2%
Identity: 13%
PDB header:unknown function
Chain: A: PDB Molecule:putative pii-like nitrogen regulatory protein;
PDBTitle: crystal structure of a duf3240 family protein (sbal_0098) from2 shewanella baltica os155 at 2.40 a resolution
Phyre2
| 16 |
|
PDB 2i3e chain A
Region: 30 - 85
Aligned: 56
Modelled: 56
Confidence: 25.5%
Identity: 14%
PDB header:hydrolase
Chain: A: PDB Molecule:g-rich;
PDBTitle: solution structure of catalytic domain of goldfish rich2 protein
Phyre2
| 17 |
|
PDB 5af2 chain C
Region: 27 - 127
Aligned: 92
Modelled: 101
Confidence: 21.2%
Identity: 17%
PDB header:hydrolase
Chain: C: PDB Molecule:vp3;
PDBTitle: crystal structure of the c-terminal 2',5'-phosphodiesterase2 domain of group a rotavirus protein vp3
Phyre2
| 18 |
|
PDB 2ilx chain A domain 1
Region: 34 - 142
Aligned: 99
Modelled: 109
Confidence: 20.5%
Identity: 22%
Fold: LigT-like
Superfamily: LigT-like
Family: 2',3'-cyclic nucleotide 3'-phosphodiesterase, catalytic domain
Phyre2
| 19 |
|
PDB 2n9z chain A
Region: 115 - 124
Aligned: 10
Modelled: 10
Confidence: 6.1%
Identity: 20%
PDB header:toxin
Chain: A: PDB Molecule:tau-theraphotoxin-hs1a;
PDBTitle: solution structure of k1 lobe of double-knot toxin
Phyre2