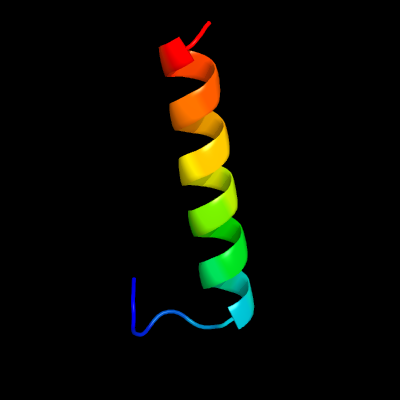
1 d2az0a1
25.6
44
Fold: ROP-likeSuperfamily: FHV B2 protein-likeFamily: FHV B2 protein-like
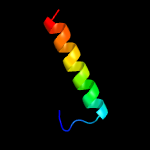
2 c2mmpA_
22.2
32
PDB header: ribosomal proteinChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of a ribosomal protein
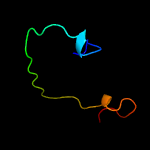
3 d1yq2a4
21.7
24
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: beta-Galactosidase, domain 5
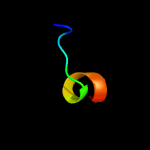
4 d1eexa_
21.3
73
Fold: TIM beta/alpha-barrelSuperfamily: Cobalamin (vitamin B12)-dependent enzymesFamily: Diol dehydratase, alpha subunit
5 d1iwpa_
20.6
73
Fold: TIM beta/alpha-barrelSuperfamily: Cobalamin (vitamin B12)-dependent enzymesFamily: Diol dehydratase, alpha subunit
6 c4gs1A_
20.1
29
PDB header: oxidoreductaseChain: A: PDB Molecule: dyp-type peroxidase;PDBTitle: crystal structure of dyp-type peroxidase from thermobifida2 cellulosilytica
7 c3bgaB_
18.5
33
PDB header: hydrolaseChain: B: PDB Molecule: beta-galactosidase;PDBTitle: crystal structure of beta-galactosidase from bacteroides2 thetaiotaomicron vpi-5482
8 c3obaA_
15.8
24
PDB header: hydrolaseChain: A: PDB Molecule: beta-galactosidase;PDBTitle: structure of the beta-galactosidase from kluyveromyces lactis
9 d1jz8a4
14.9
26
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: beta-Galactosidase, domain 5
10 c5doiF_
14.2
59
PDB header: dna binding proteinChain: F: PDB Molecule: telomerase associated protein p45;PDBTitle: crystal structure of tetrahymena p45n and p19
11 c1yq2C_
13.8
22
PDB header: hydrolaseChain: C: PDB Molecule: beta-galactosidase;PDBTitle: beta-galactosidase from arthrobacter sp. c2-2 (isoenzyme c2-2 2-1)
12 c3fubA_
12.1
19
PDB header: hormoneChain: A: PDB Molecule: gdnf family receptor alpha-1;PDBTitle: crystal structure of gdnf-gfralpha1 complex
13 c6etzA_
11.0
27
PDB header: hydrolaseChain: A: PDB Molecule: beta-galactosidase;PDBTitle: cold-adapted beta-d-galactosidase from arthrobacter sp. 32cb
14 c2v5eA_
10.7
19
PDB header: receptor/glycoprotein complexChain: A: PDB Molecule: gdnf family receptor alpha-1;PDBTitle: the structure of the gdnf:coreceptor complex: insights2 into ret signalling and heparin binding.
15 c5u1sA_
10.1
39
PDB header: hydrolaseChain: A: PDB Molecule: separin;PDBTitle: crystal structure of the saccharomyces cerevisiae separase-securin2 complex at 3.0 angstrom resolution
16 c3wknG_
7.7
63
PDB header: immune systemChain: G: PDB Molecule: affinger p17;PDBTitle: crystal structure of the artificial protein affinger p17 (af.p17)2 complexed with fc fragment of human igg
17 c3gd9A_
7.5
44
PDB header: hydrolaseChain: A: PDB Molecule: laminaripentaose-producing beta-1,3-guluasePDBTitle: crystal structure of laminaripentaose-producing beta-1,3-2 glucanase in complex with laminaritetraose
18 d2pnwa1
7.4
50
Fold: Double psi beta-barrelSuperfamily: Barwin-like endoglucanasesFamily: MLTA-like
19 c2kg4A_
7.0
17
PDB header: cell cycleChain: A: PDB Molecule: growth arrest and dna-damage-inducible protein gadd45PDBTitle: three-dimensional structure of human gadd45alpha in solution by nmr
20 c5h9xA_
6.8
67
PDB header: hydrolaseChain: A: PDB Molecule: beta-1,3-glucanase;PDBTitle: crystal structure of gh family 64 laminaripentaose-producing beta-1,3-2 glucanase from paenibacillus barengoltzii
21 c5h4eA_
not modelled
6.6
56
PDB header: hydrolaseChain: A: PDB Molecule: beta 1-3 glucanase;PDBTitle: crystal structure of a beta-1,3-glucanase domain (gh64) from2 clostridium beijerinckii
22 c2myxA_
not modelled
6.6
28
PDB header: ubiquitin-binding proteinChain: A: PDB Molecule: coupling of ubiquitin conjugation to er degradation proteinPDBTitle: structure of the cue domain of yeast cue1
23 c3m0zD_
not modelled
6.4
50
PDB header: lyaseChain: D: PDB Molecule: putative aldolase;PDBTitle: crystal structure of putative aldolase from klebsiella pneumoniae.
24 c6c3aB_
not modelled
6.3
23
PDB header: biosynthetic proteinChain: B: PDB Molecule: uncharacterized protein;PDBTitle: o2-, plp-dependent l-arginine hydroxylase rohp 4-hydroxy-2-2 ketoarginine complex
25 c3s7xC_
not modelled
6.2
22
PDB header: viral proteinChain: C: PDB Molecule: major capsid protein vp1;PDBTitle: unassembled washington university polyomavirus vp1 pentamer r198k2 mutant
26 c1zkaA_
not modelled
6.2
42
PDB header: transcriptionChain: A: PDB Molecule: transcription factor relb;PDBTitle: nf-kb relb forms an intertwined homodimer, y300s mutant
27 c5tdyC_
not modelled
6.0
41
PDB header: motor proteinChain: C: PDB Molecule: flagellar m-ring protein;PDBTitle: structure of cofolded flifc:flign complex from thermotoga maritima
28 d2g7oa1
not modelled
5.9
29
Fold: TraM-likeSuperfamily: TraM-likeFamily: TraM-like
29 c6mn8A_
not modelled
5.7
23
PDB header: ligaseChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of prolyl-trna synthetase from onchocerca volvulus2 with bound halofuginone and nucleotide
30 c3bn8A_
not modelled
5.6
22
PDB header: transport proteinChain: A: PDB Molecule: putative sterol carrier protein 2;PDBTitle: crystal structure of a putative sterol carrier protein type 2 (af1534)2 from archaeoglobus fulgidus dsm 4304 at 2.11 a resolution
31 c2dhyA_
not modelled
5.4
19
PDB header: immune systemChain: A: PDB Molecule: cue domain-containing protein 1;PDBTitle: solution structure of the cue domain in the human cue2 domain containing protein 1 (cuedc1)
32 d1g5ha1
not modelled
5.4
20
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Anticodon-binding domain of Class II aaRS
33 d2g5da1
not modelled
5.4
28
Fold: Double psi beta-barrelSuperfamily: Barwin-like endoglucanasesFamily: MLTA-like
34 c2ejsA_
not modelled
5.4
28
PDB header: ligaseChain: A: PDB Molecule: autocrine motility factor receptor, isoform 2;PDBTitle: solution structure of ruh-076, a human cue domain
35 c2ekfA_
not modelled
5.3
28
PDB header: ligaseChain: A: PDB Molecule: ancient ubiquitous protein 1;PDBTitle: solution structure of ruh-075, a human cue domain
36 c6mr1A_
not modelled
5.2
63
PDB header: protein bindingChain: A: PDB Molecule: carbon dioxide concentrating mechanism protein;PDBTitle: rbcs-like subdomain of ccmm
37 c6k1hF_
not modelled
5.1
20
PDB header: protein transportChain: F: PDB Molecule: pts system mannose-specific eiid component;PDBTitle: structure of membrane protein
38 c2prxB_
not modelled
5.0
10
PDB header: hydrolaseChain: B: PDB Molecule: thioesterase superfamily protein;PDBTitle: crystal structure of thioesterase superfamily protein (zp_00837258.1)2 from shewanella loihica pv-4 at 1.65 a resolution