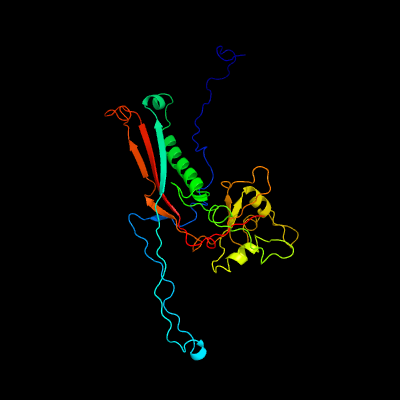
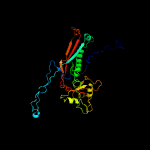
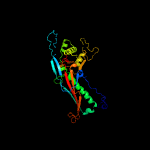
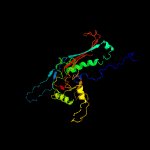
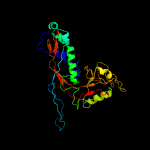
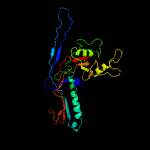
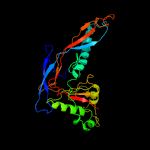
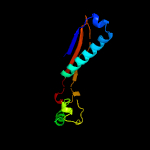
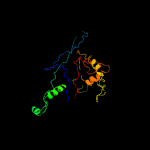
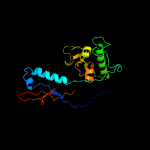
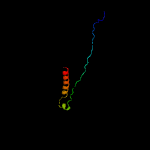
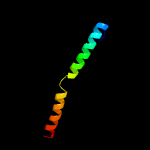
1 c5tjtA_
100.0
17
PDB header: viral proteinChain: A: PDB Molecule: major capsid protein;PDBTitle: t5 bacteriophage major capsid protein - one pb8 hexon
2 d2ft1a1
100.0
15
Fold: Major capsid protein gp5Superfamily: Major capsid protein gp5Family: Major capsid protein gp5
3 c3jb5C_
100.0
13
PDB header: virusChain: C: PDB Molecule: major capsid protein;PDBTitle: capsid structure of the propionibacterium acnes bacteriophage2 atcc_clear
4 c6b0xG_
100.0
14
PDB header: virusChain: G: PDB Molecule: major head protein;PDBTitle: capsid protein and c-terminal part of scaffolding protein in the2 staphylococcus aureus phage 80alpha procapsid
5 c1if0A_
100.0
14
PDB header: virusChain: A: PDB Molecule: protein (major capsid protein gp5);PDBTitle: pseudo-atomic model of bacteriophage hk97 procapsid2 (prohead ii)
6 c3p8qF_
100.0
15
PDB header: virusChain: F: PDB Molecule: gp5, head protein;PDBTitle: hk97 prohead i encapsidating inactive virally encoded protease
7 c3j1aE_
99.5
24
PDB header: virusChain: E: PDB Molecule: capsid protein;PDBTitle: hk97-like fold fitted into 3d reconstruction of bacteriophage cw02
8 c3j4uA_
98.7
18
PDB header: virusChain: A: PDB Molecule: major capsid protein;PDBTitle: a new topology of the hk97-like fold revealed in bordetella2 bacteriophage: non-covalent chainmail secured by jellyrolls
9 c3j7wF_
98.4
14
PDB header: virusChain: F: PDB Molecule: major capsid protein 10a;PDBTitle: capsid expansion mechanism of bacteriophage t7 revealed by multi-state2 atomic models derived from cryo-em reconstructions
10 c4bmlA_
98.2
12
PDB header: virusChain: A: PDB Molecule: major capsid protein;PDBTitle: c-alpha backbone trace of major capsid protein gp39 found in marine2 virus syn5.
11 c2e0zA_
98.1
10
PDB header: virus like particleChain: A: PDB Molecule: virus-like particle;PDBTitle: crystal structure of virus-like particle from pyrococcus2 furiosus
12 c4pt2B_
98.1
10
PDB header: virus like particleChain: B: PDB Molecule: encapsulin protein;PDBTitle: myxococcus xanthus encapsulin protein (enca)
13 c3dktD_
97.9
12
PDB header: structural protein/virus like particleChain: D: PDB Molecule: maritimacin;PDBTitle: crystal structure of thermotoga maritima encapsulin
14 c3c5bF_
97.7
16
PDB header: virusChain: F: PDB Molecule: putative uncharacterized protein;PDBTitle: de novo model of bacteriophage epsilon 15 major capsid protein gp7
15 c4an5F_
97.2
12
PDB header: virusChain: F: PDB Molecule: coat protein;PDBTitle: capsid structure and its stability at the late stages of bacteriophage2 spp1 assembly
16 c2xd8F_
95.1
13
PDB header: virusChain: F: PDB Molecule: t7-like capsid protein;PDBTitle: capsid structure of the infectious prochlorococcus cyanophage p-ssp7
17 c1yueA_
91.7
13
PDB header: viral proteinChain: A: PDB Molecule: head vertex protein gp24;PDBTitle: bacteriophage t4 capsid vertex protein gp24
18 c5vf3H_
87.7
20
PDB header: virusChain: H: PDB Molecule: major capsid protein;PDBTitle: bacteriophage t4 isometric capsid
19 c1j1eB_
60.6
20
PDB header: contractile proteinChain: B: PDB Molecule: troponin t;PDBTitle: crystal structure of the 52kda domain of human cardiac2 troponin in the ca2+ saturated form
20 c1j1dF_
37.5
17
PDB header: contractile proteinChain: F: PDB Molecule: troponin i;PDBTitle: crystal structure of the 46kda domain of human cardiac2 troponin in the ca2+ saturated form
21 c5liiP_
not modelled
34.2
12
PDB header: virus like particleChain: P: PDB Molecule: major capsid protein;PDBTitle: bacteriophage phi812k1-420 major capsid protein
22 c5eonA_
not modelled
28.6
18
PDB header: de novo proteinChain: A: PDB Molecule: acc-hex;PDBTitle: crystal structure of a de novo antiparallel coiled-coil hexamer - acc-2 hex
23 c5eonB_
not modelled
28.6
18
PDB header: de novo proteinChain: B: PDB Molecule: acc-hex;PDBTitle: crystal structure of a de novo antiparallel coiled-coil hexamer - acc-2 hex
24 c5eonC_
not modelled
28.6
18
PDB header: de novo proteinChain: C: PDB Molecule: acc-hex;PDBTitle: crystal structure of a de novo antiparallel coiled-coil hexamer - acc-2 hex
25 c1yv0T_
not modelled
28.0
21
PDB header: contractile proteinChain: T: PDB Molecule: troponin t, fast skeletal muscle isoforms;PDBTitle: crystal structure of skeletal muscle troponin in the ca2+-free state
26 c1j1eF_
not modelled
23.0
17
PDB header: contractile proteinChain: F: PDB Molecule: troponin i;PDBTitle: crystal structure of the 52kda domain of human cardiac2 troponin in the ca2+ saturated form
27 c5l35D_
not modelled
17.6
10
PDB header: virusChain: D: PDB Molecule: gene 5 protein;PDBTitle: cryo-em structure of bacteriophage sf6 at 2.9 angstrom resolution
28 c1j1eC_
not modelled
16.7
17
PDB header: contractile proteinChain: C: PDB Molecule: troponin i;PDBTitle: crystal structure of the 52kda domain of human cardiac2 troponin in the ca2+ saturated form
29 d1vqov1
not modelled
16.2
14
Fold: Long alpha-hairpinSuperfamily: Ribosomal protein L29 (L29p)Family: Ribosomal protein L29 (L29p)
30 c3izgG_
not modelled
15.9
19
PDB header: virusChain: G: PDB Molecule: major capsid protein 10a;PDBTitle: bacteriophage t7 prohead shell em-derived atomic model
31 d1jeia_
not modelled
12.1
22
Fold: LEM/SAP HeH motifSuperfamily: LEM domainFamily: LEM domain
32 c4l6rA_
not modelled
11.8
15
PDB header: membrane proteinChain: A: PDB Molecule: soluble cytochrome b562 and glucagon receptor chimera;PDBTitle: structure of the class b human glucagon g protein coupled receptor
33 c3uotB_
not modelled
11.7
8
PDB header: cell cycleChain: B: PDB Molecule: mediator of dna damage checkpoint protein 1;PDBTitle: crystal structure of mdc1 fha domain in complex with a phosphorylated2 peptide from the mdc1 n-terminus
34 c3d55A_
not modelled
9.8
27
PDB header: toxin inhibitorChain: A: PDB Molecule: uncharacterized protein rv3357/mt3465;PDBTitle: crystal structure of m. tuberculosis yefm antitoxin
35 c4bjsC_
not modelled
9.7
35
PDB header: cell cycleChain: C: PDB Molecule: telomere length regulator protein rif1;PDBTitle: crystal structure of the rif1 c-terminal domain (rif1-ctd)2 from saccharomyces cerevisiae
36 d1s35a1
not modelled
9.3
13
Fold: Spectrin repeat-likeSuperfamily: Spectrin repeatFamily: Spectrin repeat
37 c3jcuw_
not modelled
9.1
33
PDB header: membrane proteinChain: W: PDB Molecule: photosystem ii reaction center w protein, chloroplastic;PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
38 c3bqwA_
not modelled
8.5
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative capsid protein of prophage;PDBTitle: crystal structure of the putative capsid protein of prophage (e.coli2 cft073)
39 d2hgq11
not modelled
8.0
14
Fold: Long alpha-hairpinSuperfamily: Ribosomal protein L29 (L29p)Family: Ribosomal protein L29 (L29p)
40 c2myvA_
not modelled
7.8
6
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of m. oryzae protein avr1-co39
41 c2krxA_
not modelled
7.7
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: asl3597 protein;PDBTitle: solution nmr structure of asl3597 from nostoc sp. pcc7120. northeast2 structural genomics consortium target id nsr244.
42 c3jtzA_
not modelled
7.6
29
PDB header: dna binding proteinChain: A: PDB Molecule: integrase;PDBTitle: structure of the arm-type binding domain of hpi integrase
43 c1ytzI_
not modelled
7.1
16
PDB header: contractile proteinChain: I: PDB Molecule: troponin i;PDBTitle: crystal structure of skeletal muscle troponin in the ca2+-2 activated state
44 d1tdha3
not modelled
7.0
13
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins
45 d1gjja2
not modelled
6.8
18
Fold: LEM/SAP HeH motifSuperfamily: LEM domainFamily: LEM domain
46 d2ho2a1
not modelled
6.6
8
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain
47 d1dd3a1
not modelled
6.4
21
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domainSuperfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domainFamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain
48 d2vo1a1
not modelled
6.3
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like
49 c3bboZ_
not modelled
6.2
14
PDB header: ribosomeChain: Z: PDB Molecule: ribosomal protein l29;PDBTitle: homology model for the spinach chloroplast 50s subunit fitted to 9.4a2 cryo-em map of the 70s chlororibosome
50 c3m8jA_
not modelled
5.8
8
PDB header: transcriptionChain: A: PDB Molecule: focb protein;PDBTitle: crystal structure of e.coli focb at 1.4 a resolution
51 c5mlcZ_
not modelled
5.6
14
PDB header: ribosomeChain: Z: PDB Molecule: 50s ribosomal protein l29, chloroplastic;PDBTitle: cryo-em structure of the spinach chloroplast ribosome reveals the2 location of plastid-specific ribosomal proteins and extensions
52 c6cc4A_
not modelled
5.6
8
PDB header: transport proteinChain: A: PDB Molecule: soluble cytochrome b562, lipid ii flippase murj chimera;PDBTitle: structure of murj from escherichia coli
53 d1h9fa_
not modelled
5.5
14
Fold: LEM/SAP HeH motifSuperfamily: LEM domainFamily: LEM domain
54 c5cwtC_
not modelled
5.5
24
PDB header: transport proteinChain: C: PDB Molecule: nucleoporin nup57;PDBTitle: crystal structure of chaetomium thermophilum nup57