| 1 |
|
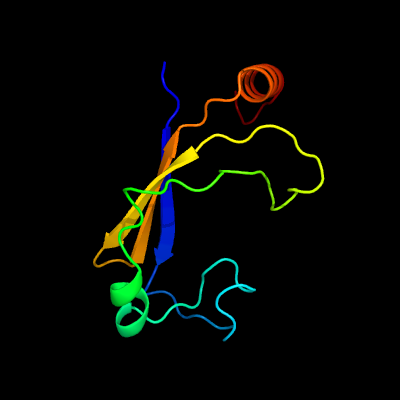
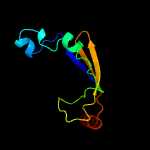
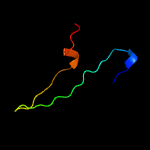
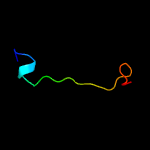
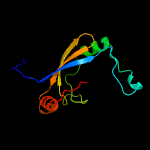
PDB 2pbk chain A domain 1
Region: 8 - 105
Aligned: 89
Modelled: 98
Confidence: 95.6%
Identity: 18%
Fold: Herpes virus serine proteinase, assemblin
Superfamily: Herpes virus serine proteinase, assemblin
Family: Herpes virus serine proteinase, assemblin
Phyre2
| 2 |
|
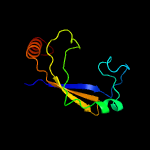
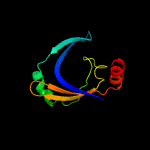
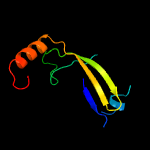
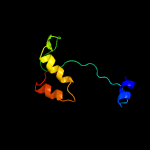
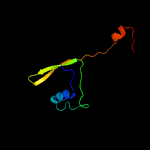
PDB 1vzv chain A
Region: 8 - 105
Aligned: 87
Modelled: 98
Confidence: 94.6%
Identity: 15%
Fold: Herpes virus serine proteinase, assemblin
Superfamily: Herpes virus serine proteinase, assemblin
Family: Herpes virus serine proteinase, assemblin
Phyre2
| 3 |
|
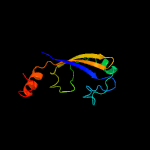
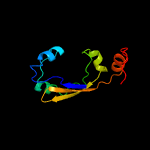
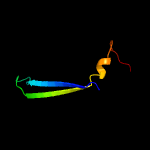
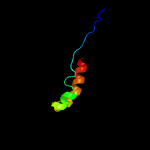
PDB 1o6e chain A
Region: 12 - 86
Aligned: 66
Modelled: 75
Confidence: 94.4%
Identity: 24%
Fold: Herpes virus serine proteinase, assemblin
Superfamily: Herpes virus serine proteinase, assemblin
Family: Herpes virus serine proteinase, assemblin
Phyre2
| 4 |
|
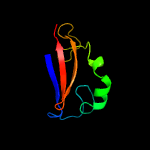
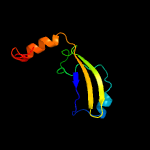
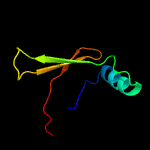
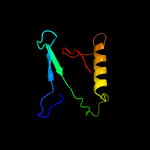
PDB 1ieg chain A
Region: 12 - 98
Aligned: 74
Modelled: 87
Confidence: 93.3%
Identity: 16%
Fold: Herpes virus serine proteinase, assemblin
Superfamily: Herpes virus serine proteinase, assemblin
Family: Herpes virus serine proteinase, assemblin
Phyre2
| 5 |
|
PDB 3njq chain B
Region: 9 - 98
Aligned: 80
Modelled: 90
Confidence: 92.6%
Identity: 19%
PDB header:viral protein/inhibitor
Chain: B: PDB Molecule:orf 17;
PDBTitle: crystal structure of kaposi's sarcoma-associated herpesvirus protease2 in complex with dimer disruptor
Phyre2
| 6 |
|
PDB 1at3 chain A
Region: 12 - 103
Aligned: 82
Modelled: 92
Confidence: 92.0%
Identity: 23%
Fold: Herpes virus serine proteinase, assemblin
Superfamily: Herpes virus serine proteinase, assemblin
Family: Herpes virus serine proteinase, assemblin
Phyre2
| 7 |
|
PDB 4cx8 chain B
Region: 12 - 107
Aligned: 78
Modelled: 81
Confidence: 91.6%
Identity: 17%
PDB header:viral protein
Chain: B: PDB Molecule:pseudorabies virus protease;
PDBTitle: monomeric pseudorabies virus protease pul26n at 2.5 a resolution
Phyre2
| 8 |
|
PDB 2ed6 chain A domain 1
Region: 3 - 39
Aligned: 34
Modelled: 37
Confidence: 41.7%
Identity: 24%
Fold: WSSV envelope protein-like
Superfamily: WSSV envelope protein-like
Family: WSSV envelope protein-like
Phyre2
| 9 |
|
PDB 1jq6 chain A
Region: 12 - 105
Aligned: 71
Modelled: 74
Confidence: 37.8%
Identity: 24%
Fold: Herpes virus serine proteinase, assemblin
Superfamily: Herpes virus serine proteinase, assemblin
Family: Herpes virus serine proteinase, assemblin
Phyre2
| 10 |
|
PDB 3cax chain A
Region: 128 - 170
Aligned: 42
Modelled: 43
Confidence: 30.2%
Identity: 14%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein pf0695;
PDBTitle: crystal structure of uncharacterized protein pf0695
Phyre2
| 11 |
|
PDB 4oy2 chain D
Region: 81 - 139
Aligned: 53
Modelled: 59
Confidence: 18.7%
Identity: 11%
PDB header:transcription
Chain: D: PDB Molecule:transcription initiation factor tfiid subunit 7;
PDBTitle: crystal structure of taf1-taf7, a tfiid subcomplex
Phyre2
| 12 |
|
PDB 1a8d chain A domain 2
Region: 98 - 120
Aligned: 22
Modelled: 23
Confidence: 14.7%
Identity: 18%
Fold: beta-Trefoil
Superfamily: STI-like
Family: Clostridium neurotoxins, C-terminal domain
Phyre2
| 13 |
|
PDB 4x8r chain B
Region: 41 - 106
Aligned: 64
Modelled: 66
Confidence: 8.4%
Identity: 14%
PDB header:solute-binding protein
Chain: B: PDB Molecule:trap dicarboxylate transporter, dctp subunit;
PDBTitle: crystal structure of a trap periplasmic solute binding protein from2 rhodobacter sphaeroides (rsph17029_2138, target efi-510205) with3 bound glucuronate
Phyre2
| 14 |
|
PDB 1tdh chain A domain 3
Region: 1 - 12
Aligned: 12
Modelled: 12
Confidence: 6.8%
Identity: 42%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins
Phyre2
| 15 |
|
PDB 5zdc chain L
Region: 128 - 168
Aligned: 40
Modelled: 41
Confidence: 6.3%
Identity: 15%
PDB header:hydrolase
Chain: L: PDB Molecule:poly adp-ribose glycohydrolase;
PDBTitle: crystal structure of poly(adp-ribose) glycohydrolase (parg) from2 deinococcus radiodurans in complex with adp-ribose (p32)
Phyre2
| 16 |
|
PDB 4zlp chain B
Region: 45 - 116
Aligned: 66
Modelled: 72
Confidence: 5.8%
Identity: 12%
PDB header:transcription
Chain: B: PDB Molecule:neurogenic locus notch homolog protein 3;
PDBTitle: crystal structure of notch3 negative regulatory region
Phyre2
| 17 |
|
PDB 5jbl chain A
Region: 1 - 107
Aligned: 100
Modelled: 107
Confidence: 5.6%
Identity: 13%
PDB header:hydrolase
Chain: A: PDB Molecule:prohead core protein protease;
PDBTitle: structure of the bacteriophage t4 capsid assembly protease, gp21.
Phyre2
| 18 |
|
PDB 1ckq chain A
Region: 32 - 105
Aligned: 73
Modelled: 74
Confidence: 5.5%
Identity: 12%
Fold: Restriction endonuclease-like
Superfamily: Restriction endonuclease-like
Family: Restriction endonuclease EcoRI
Phyre2