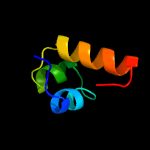
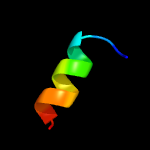
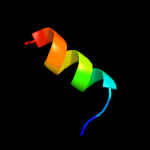
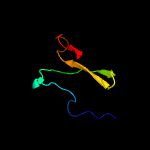
1 c1tr8A_
92.2
40
PDB header: chaperoneChain: A: PDB Molecule: conserved protein (mth177);PDBTitle: crystal structure of archaeal nascent polypeptide-associated complex2 (aenac)
2 c6riqP_
73.5
31
PDB header: protein fibrilChain: P: PDB Molecule: minc;PDBTitle: mincd filament from pseudomonas aeruginosa
3 c4v02C_
70.8
31
PDB header: cell cycleChain: C: PDB Molecule: probable septum site-determining protein minc;PDBTitle: minc:mind cell division protein complex, aquifex aeolicus
4 c5xdmA_
64.4
29
PDB header: cell cycleChain: A: PDB Molecule: septum site-determining protein minc;PDBTitle: structure of the c-terminal domain of e. coli minc at 3.0 angstrom2 resolution
5 d1hf2a1
64.3
26
Fold: Single-stranded right-handed beta-helixSuperfamily: Cell-division inhibitor MinC, C-terminal domainFamily: Cell-division inhibitor MinC, C-terminal domain
6 c5fjaQ_
59.9
69
PDB header: transcriptionChain: Q: PDB Molecule: dna-directed rna polymerase iii subunit rpc7;PDBTitle: cryo-em structure of yeast rna polymerase iii at 4.7 a
7 c6cnbQ_
56.6
69
PDB header: transcription/dnaChain: Q: PDB Molecule: dna-directed rna polymerase iii subunit rpc7,dna-directedPDBTitle: yeast rna polymerase iii initial transcribing complex
8 c6cncQ_
56.6
69
PDB header: transcription/dnaChain: Q: PDB Molecule: dna-directed rna polymerase iii subunit rpc7,dna-directedPDBTitle: yeast rna polymerase iii open complex
9 c6cndQ_
56.4
69
PDB header: transcription/dnaChain: Q: PDB Molecule: dna-directed rna polymerase iii subunit rpc7,dna-directedPDBTitle: yeast rna polymerase iii natural open complex (noc)
10 c6cnfQ_
56.4
69
PDB header: transcription/dnaChain: Q: PDB Molecule: dna-directed rna polymerase iii subunit rpc7,dna-directedPDBTitle: yeast rna polymerase iii elongation complex
11 c5fj8Q_
55.5
69
PDB header: transcriptionChain: Q: PDB Molecule: dna-directed rna polymerase iii subunit rpc7;PDBTitle: cryo-em structure of yeast rna polymerase iii elongation complex at 3.2 9 a
12 c1hf2A_
53.1
26
PDB header: cell division proteinChain: A: PDB Molecule: septum site-determining protein minc;PDBTitle: crystal structure of the bacterial cell-division inhibitor minc from2 t. maritima
13 c5fj9Q_
52.6
69
PDB header: transcriptionChain: Q: PDB Molecule: dna-directed rna polymerase iii subunit rpc7;PDBTitle: cryo-em structure of yeast apo rna polymerase iii at 4.6 a
14 d1v92a_
44.9
32
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: TAP-C domain-like
15 d1ou8a_
43.8
29
Fold: SspB-likeSuperfamily: SspB-likeFamily: Stringent starvation protein B, SspB
16 d1k78a1
41.7
30
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain
17 d1msza_
41.4
36
Fold: IF3-likeSuperfamily: R3H domainFamily: R3H domain
18 c1mszA_
41.4
36
PDB header: dna binding proteinChain: A: PDB Molecule: dna-binding protein smubp-2;PDBTitle: solution structure of the r3h domain from human smubp-2
19 c3akgA_
37.4
30
PDB header: hydrolaseChain: A: PDB Molecule: putative secreted alpha l-arabinofuranosidase ii;PDBTitle: crystal structure of exo-1,5-alpha-l-arabinofuranosidase complexed2 with alpha-1,5-l-arabinofuranobiose
20 c2dzlA_
30.0
42
PDB header: structural genomics unknown functionChain: A: PDB Molecule: protein fam100b;PDBTitle: solution structure of the uba domain in human protein2 fam100b
21 c5zjlA_
not modelled
29.1
58
PDB header: allergenChain: A: PDB Molecule: der f 23 allergen;PDBTitle: crystal structure of the dust mite allergen der f 23 from2 dermatophagoides farinae
22 d1pdnc_
not modelled
28.8
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain
23 d1ou9a_
not modelled
26.5
29
Fold: SspB-likeSuperfamily: SspB-likeFamily: Stringent starvation protein B, SspB
24 d1yfna1
not modelled
25.3
24
Fold: SspB-likeSuperfamily: SspB-likeFamily: Stringent starvation protein B, SspB
25 c6paxA_
not modelled
24.1
26
PDB header: gene regulation/dnaChain: A: PDB Molecule: homeobox protein pax-6;PDBTitle: crystal structure of the human pax-6 paired domain-dna2 complex reveals a general model for pax protein-dna3 interactions
26 d2csba1
not modelled
24.0
64
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Topoisomerase V repeat domain
27 d1gg4a4
not modelled
20.8
19
Fold: Ribokinase-likeSuperfamily: MurD-like peptide ligases, catalytic domainFamily: MurCDEF
28 d2euca1
not modelled
20.6
31
Fold: YfmB-likeSuperfamily: YfmB-likeFamily: YfmB-like
29 d1zszc1
not modelled
20.1
26
Fold: SspB-likeSuperfamily: SspB-likeFamily: Stringent starvation protein B, SspB
30 d6paxa1
not modelled
18.3
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain
31 c5gkeB_
not modelled
17.8
19
PDB header: hydrolase/dnaChain: B: PDB Molecule: endonuclease endoms;PDBTitle: structure of endoms-dsdna1 complex
32 c2cpmA_
not modelled
17.3
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: sperm-associated antigen 7;PDBTitle: solution structure of the r3h domain of human sperm-2 associated antigen 7
33 d1ko7a1
not modelled
16.5
18
Fold: MurF and HprK N-domain-likeSuperfamily: HprK N-terminal domain-likeFamily: HPr kinase/phoshatase HprK N-terminal domain
34 c3csqC_
not modelled
15.9
22
PDB header: hydrolaseChain: C: PDB Molecule: morphogenesis protein 1;PDBTitle: crystal and cryoem structural studies of a cell wall2 degrading enzyme in the bacteriophage phi29 tail
35 c5ho0A_
not modelled
15.2
21
PDB header: hydrolaseChain: A: PDB Molecule: extracellular arabinanase;PDBTitle: crystal structure of abna (closed conformation), a gh43 extracellular2 arabinanase from geobacillus stearothermophilus
36 d2hyec3
not modelled
14.3
15
Fold: Cullin homology domainSuperfamily: Cullin homology domainFamily: Cullin homology domain
37 c5u0pT_
not modelled
14.2
34
PDB header: transcriptionChain: T: PDB Molecule: mediator complex subunit 20;PDBTitle: cryo-em structure of the transcriptional mediator
38 d1omza_
not modelled
13.6
7
Fold: Nucleotide-diphospho-sugar transferasesSuperfamily: Nucleotide-diphospho-sugar transferasesFamily: Exostosin
39 c3ixtC_
not modelled
13.5
60
PDB header: immune systemChain: C: PDB Molecule: fusion glycoprotein f1;PDBTitle: crystal structure of motavizumab fab bound to peptide epitope
40 c3ixtP_
not modelled
13.5
60
PDB header: immune systemChain: P: PDB Molecule: fusion glycoprotein f1;PDBTitle: crystal structure of motavizumab fab bound to peptide epitope
41 c3kstA_
not modelled
12.6
17
PDB header: hydrolaseChain: A: PDB Molecule: endo-1,4-beta-xylanase;PDBTitle: crystal structure of endo-1,4-beta-xylanase (np_811807.1) from2 bacteroides thetaiotaomicron vpi-5482 at 1.70 a resolution
42 d1tiha_
not modelled
12.0
33
Fold: Plant proteinase inhibitorsSuperfamily: Plant proteinase inhibitorsFamily: Plant proteinase inhibitors
43 c1u78A_
not modelled
11.2
16
PDB header: dna binding protein/dnaChain: A: PDB Molecule: transposable element tc3 transposase;PDBTitle: structure of the bipartite dna-binding domain of tc32 transposase bound to transposon dna
44 c1omxB_
not modelled
11.2
6
PDB header: transferaseChain: B: PDB Molecule: alpha-1,4-n-acetylhexosaminyltransferase extl2;PDBTitle: crystal structure of mouse alpha-1,4-n-acetylhexosaminyltransferase2 (extl2)
45 d1pjua2
not modelled
11.1
33
Fold: Plant proteinase inhibitorsSuperfamily: Plant proteinase inhibitorsFamily: Plant proteinase inhibitors
46 c2opwA_
not modelled
10.9
44
PDB header: oxidoreductaseChain: A: PDB Molecule: phyhd1 protein;PDBTitle: crystal structure of human phytanoyl-coa dioxygenase phyhd1 (apo)
47 d1whra_
not modelled
10.6
36
Fold: IF3-likeSuperfamily: R3H domainFamily: R3H domain
48 c1fvyA_
not modelled
10.1
75
PDB header: hormone/growth factorChain: A: PDB Molecule: parathyroid hormone;PDBTitle: solution structure of the osteogenic 1-31 fragment of the2 human parathyroid hormone
49 c6eu1Q_
not modelled
10.1
50
PDB header: transcriptionChain: Q: PDB Molecule: dna-directed rna polymerase iii subunit rpc7;PDBTitle: rna polymerase iii - open dna complex (oc-pol3)
50 c2k27A_
not modelled
9.8
28
PDB header: transcription regulatorChain: A: PDB Molecule: paired box protein pax-8;PDBTitle: solution structure of human pax8 paired box domain
51 c3bmzA_
not modelled
9.5
27
PDB header: biosynthetic proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: violacein biosynthetic enzyme vioe
52 c3zljC_
not modelled
9.1
55
PDB header: dna binding protein/dnaChain: C: PDB Molecule: dna mismatch repair protein muts;PDBTitle: crystal structure of full-length e.coli dna mismatch repair protein2 muts d835r mutant in complex with gt mismatched dna
53 c6cauA_
not modelled
9.0
29
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate--l-alanine ligase;PDBTitle: udp-n-acetylmuramate--alanine ligase from acinetobacter baumannii2 ab5075-uw with amppnp
54 d1ayaa_
not modelled
9.0
22
Fold: SH2-likeSuperfamily: SH2 domainFamily: SH2 domain
55 c5xtaC_
not modelled
8.7
21
PDB header: oxidoreductaseChain: C: PDB Molecule: virk protein;PDBTitle: crystal structure of lpg1832, a virk family protein from legionella2 pneumophila
56 c2ge9A_
not modelled
8.4
26
PDB header: transferaseChain: A: PDB Molecule: tyrosine-protein kinase btk;PDBTitle: solution structures of the sh2 domain of bruton's tyrosine2 kinase
57 c1wn4A_
not modelled
8.3
67
PDB header: plant proteinChain: A: PDB Molecule: vontr protein;PDBTitle: nmr structure of vontr
58 c3zljD_
not modelled
8.2
55
PDB header: dna binding protein/dnaChain: D: PDB Molecule: dna mismatch repair protein muts;PDBTitle: crystal structure of full-length e.coli dna mismatch repair protein2 muts d835r mutant in complex with gt mismatched dna
59 c3tr7A_
not modelled
8.2
32
PDB header: hydrolaseChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: structure of a uracil-dna glycosylase (ung) from coxiella burnetii
60 c3lv4B_
not modelled
8.1
26
PDB header: hydrolaseChain: B: PDB Molecule: glycoside hydrolase yxia;PDBTitle: crystal structure of the glycoside hydrolase, family 43 yxia protein2 from bacillus licheniformis. northeast structural genomics consortium3 target bir14.
61 d1e8ca3
not modelled
7.9
6
Fold: Ribokinase-likeSuperfamily: MurD-like peptide ligases, catalytic domainFamily: MurCDEF
62 d1moua_
not modelled
7.8
32
Fold: GFP-likeSuperfamily: GFP-likeFamily: Fluorescent proteins
63 d4sgbi_
not modelled
7.7
21
Fold: Plant proteinase inhibitorsSuperfamily: Plant proteinase inhibitorsFamily: Plant proteinase inhibitors
64 c5erlD_
not modelled
7.7
36
PDB header: isomeraseChain: D: PDB Molecule: snon,snon;PDBTitle: crystal structure of the epimerase snon in complex with ni2+,2 succinate and nogalamycin ro
65 c4ae4B_
not modelled
7.6
35
PDB header: protein transportChain: B: PDB Molecule: ubiquitin-associated protein 1;PDBTitle: the ubap1 subunit of escrt-i interacts with ubiquitin via a novel2 souba domain
66 d1s6la1
not modelled
7.5
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MerB N-terminal domain-like
67 c1x6cA_
not modelled
7.4
23
PDB header: signaling proteinChain: A: PDB Molecule: tyrosine-protein phosphatase, non-receptor typePDBTitle: solution structures of the sh2 domain of human protein-2 tyrosine phosphatase shp-1
68 c5gtuB_
not modelled
7.3
44
PDB header: hydrolaseChain: B: PDB Molecule: tbc1 domain family member 5;PDBTitle: structural and mechanistic insights into regulation of the retromer2 coat by tbc1d5
69 c4zonB_
not modelled
7.3
31
PDB header: oxidoreductase/oxidoreductase inhibitorChain: B: PDB Molecule: verruculogen synthase;PDBTitle: structure of ftmox1 with fumitremorgen b complex
70 d1fyba1
not modelled
7.1
36
Fold: Plant proteinase inhibitorsSuperfamily: Plant proteinase inhibitorsFamily: Plant proteinase inhibitors
71 d1z67a1
not modelled
7.0
16
Fold: YidB-likeSuperfamily: YidB-likeFamily: YidB-like
72 d1ldja3
not modelled
6.9
24
Fold: Cullin homology domainSuperfamily: Cullin homology domainFamily: Cullin homology domain
73 c5n6yD_
not modelled
6.8
26
PDB header: oxidoreductaseChain: D: PDB Molecule: nitrogenase vanadium-iron protein alpha chain;PDBTitle: azotobacter vinelandii vanadium nitrogenase
74 d1t1ea2
not modelled
6.8
17
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Subtilase propeptides/inhibitors
75 c5ncrA_
not modelled
6.7
30
PDB header: hydrolaseChain: A: PDB Molecule: tyrosine phosphatase;PDBTitle: oh1 from the orf virus: a tyrosine phosphatase that displays distinct2 structural features and triple substrate specificity
76 c5vvwA_
not modelled
6.6
29
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate--l-alanine ligase;PDBTitle: structure of murc from pseudomonas aeruginosa
77 c5m0tA_
not modelled
6.4
19
PDB header: oxidoreductaseChain: A: PDB Molecule: alpha-ketoglutarate-dependent non-heme iron oxygenase eash;PDBTitle: alpha-ketoglutarate-dependent non-heme iron oxygenase eash
78 c3eagA_
not modelled
6.3
29
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate:l-alanyl-gamma-d-glutamyl-meso-PDBTitle: the crystal structure of udp-n-acetylmuramate:l-alanyl-gamma-d-2 glutamyl-meso-diaminopimelate ligase (mpl) from neisseria3 meningitides
79 c1j6uA_
not modelled
6.3
29
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate-alanine ligase murc;PDBTitle: crystal structure of udp-n-acetylmuramate-alanine ligase murc (tm0231)2 from thermotoga maritima at 2.3 a resolution
80 c2booA_
not modelled
6.2
29
PDB header: hydrolaseChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: the crystal structure of uracil-dna n-glycosylase (ung) from2 deinococcus radiodurans.
81 c2mtpB_
not modelled
6.2
46
PDB header: protein binding/cell adhesionChain: B: PDB Molecule: integrin alpha-iib;PDBTitle: the structure of filamin repeat 21 bound to integrin
82 d1oyvi_
not modelled
6.2
36
Fold: Plant proteinase inhibitorsSuperfamily: Plant proteinase inhibitorsFamily: Plant proteinase inhibitors
83 d1i3za_
not modelled
6.1
16
Fold: SH2-likeSuperfamily: SH2 domainFamily: SH2 domain
84 c2lseA_
not modelled
6.1
34
PDB header: de novo proteinChain: A: PDB Molecule: four helix bundle protein;PDBTitle: solution nmr structure of de novo designed four helix bundle protein,2 northeast structural genomics consortium (nesg) target or188
85 c4xc9B_
not modelled
6.1
38
PDB header: oxidoreductaseChain: B: PDB Molecule: oxidase/hydroxylase;PDBTitle: crystal structure of apo hygx from streptomyces hygroscopicus
86 c6reqB_
not modelled
6.1
19
PDB header: isomeraseChain: B: PDB Molecule: protein (methylmalonyl-coa mutase);PDBTitle: methylmalonyl-coa mutase, 3-carboxypropyl-coa inhibitor complex
87 c2z5bB_
not modelled
5.9
56
PDB header: chaperoneChain: B: PDB Molecule: uncharacterized protein ylr021w;PDBTitle: crystal structure of a novel chaperone complex for yeast2 20s proteasome assembly
88 c3m6zA_
not modelled
5.9
64
PDB header: isomeraseChain: A: PDB Molecule: topoisomerase v;PDBTitle: crystal structure of an n-terminal 44 kda fragment of topoisomerase v2 in the presence of guanidium hydrochloride
89 c5ermA_
not modelled
5.8
24
PDB header: lyaseChain: A: PDB Molecule: fusicoccadiene synthase;PDBTitle: crystal structure of cyclization domain of phomopsis amygdali2 fusicoccadiene synthase complexed with magnesium ions and pamidronate
90 d1gyxa_
not modelled
5.8
7
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: 4-oxalocrotonate tautomerase-like
91 c3mazA_
not modelled
5.8
38
PDB header: signaling proteinChain: A: PDB Molecule: signal-transducing adaptor protein 1;PDBTitle: crystal structure of the human brdg1/stap-1 sh2 domain in complex with2 the ntal ptyr136 peptide
92 d1pjua1
not modelled
5.7
21
Fold: Plant proteinase inhibitorsSuperfamily: Plant proteinase inhibitorsFamily: Plant proteinase inhibitors
93 c2ci8A_
not modelled
5.7
20
PDB header: translationChain: A: PDB Molecule: cytoplasmic protein nck1;PDBTitle: sh2 domain of human nck1 adaptor protein - uncomplexed
94 c4xbzB_
not modelled
5.6
38
PDB header: oxidoreductaseChain: B: PDB Molecule: evdo1;PDBTitle: crystal structure of evdo1 from micromonospora carbonacea var.2 aurantiaca
95 c1m8oA_
not modelled
5.6
46
PDB header: membrane proteinChain: A: PDB Molecule: platelet integrin alfaiib subunit: cytoplasmicPDBTitle: platelet integrin alfaiib-beta3 cytoplasmic domain
96 c1s4wA_
not modelled
5.6
46
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: nmr structure of the cytoplasmic domain of integrin aiib in2 dpc micelles
97 c1dpkA_
not modelled
5.6
46
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib subunit;PDBTitle: solution structure of the cytoplasmic domain of the2 integrin alpha-iib subunit
98 c4jlrS_
not modelled
5.6
60
PDB header: immune systemChain: S: PDB Molecule: rsv_1isea designed scaffold;PDBTitle: crystal structure of a designed respiratory syncytial virus immunogen2 in complex with motavizumab
99 c5zm4B_
not modelled
5.5
25
PDB header: oxidoreductaseChain: B: PDB Molecule: dioxygenase anda;PDBTitle: fe(ii)/(alpha)ketoglutarate-dependent dioxygenase anda with2 preandiloid c