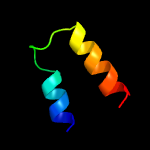
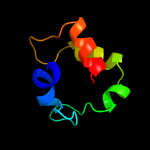
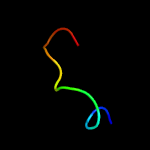
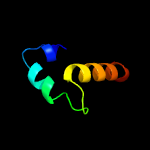
1 c1gw4A_
72.3
55
PDB header: high density lipoproteinsChain: A: PDB Molecule: apoa-i;PDBTitle: the helix-hinge-helix structural motif in human2 apolipoprotein a-i determined by nmr spectroscopy, 13 structure
2 d1u8va1
57.0
30
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: Medium chain acyl-CoA dehydrogenase-like, C-terminal domain
3 c4oo2D_
50.1
20
PDB header: oxidoreductaseChain: D: PDB Molecule: chlorophenol-4-monooxygenase;PDBTitle: streptomyces globisporus c-1027 fad dependent (s)-3-chloro-β-2 tyrosine-s-sgcc2 c-5 hydroxylase sgcc apo form
4 c3hwcD_
34.5
9
PDB header: oxidoreductaseChain: D: PDB Molecule: chlorophenol-4-monooxygenase component 2;PDBTitle: crystal structure of chlorophenol 4-monooxygenase (tftd) of2 burkholderia cepacia ac1100
5 c6eb0A_
33.4
22
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxyphenylacetate 3-monooxygenase, oxygenase subunit;PDBTitle: structure of 4-hydroxyphenylacetate 3-monooxygenase (hpab), oxygenase2 component from escherichia coli
6 d1mtyb_
31.6
26
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ribonucleotide reductase-like
7 d2q22a1
31.3
18
Fold: Ava3019-likeSuperfamily: Ava3019-likeFamily: Ava3019-like
8 c1odrA_
30.3
40
PDB header: lipid transportChain: A: PDB Molecule: apoa-i peptide;PDBTitle: peptide of human apoa-i residues 166-185. nmr, 5 structures2 at ph 6.0, 37 degrees celsius and peptide:dpc mole ratio3 of 1:40
9 c1odpA_
30.3
40
PDB header: lipid transportChain: A: PDB Molecule: apoa-i peptide;PDBTitle: peptide of human apoa-i residues 166-185. nmr, 5 structures2 at ph 6.6, 37 degrees celsius and peptide:sds mole ratio3 of 1:40
10 c1odqA_
30.3
40
PDB header: lipid transportChain: A: PDB Molecule: apoa-i peptide;PDBTitle: peptide of human apoa-i residues 166-185. nmr, 5 structures2 at ph 3.7, 37 degrees celsius and peptide:sds mole ratio3 of 1:40
11 c1u8vA_
26.1
32
PDB header: lyase, isomeraseChain: A: PDB Molecule: gamma-aminobutyrate metabolism dehydratase/isomerase;PDBTitle: crystal structure of 4-hydroxybutyryl-coa dehydratase from clostridium2 aminobutyricum: radical catalysis involving a [4fe-4s] cluster and3 flavin
12 c6ny5A_
25.3
14
PDB header: rna binding protein/rnaChain: A: PDB Molecule: pumilio domain-containing protein c56f2.08c;PDBTitle: crystal structure of the pum-hd domain of s. pombe puf1 in complex2 with rna
13 c4g5eD_
17.0
10
PDB header: oxidoreductaseChain: D: PDB Molecule: 2,4,6-trichlorophenol 4-monooxygenase;PDBTitle: 2,4,6-trichlorophenol 4-monooxygenase
14 d1hj3a1
16.2
20
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase
15 d1rbli_
16.1
50
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
16 c2kz5A_
14.7
53
PDB header: transcriptionChain: A: PDB Molecule: transcription factor nf-e2 45 kda subunit;PDBTitle: solution nmr structure of transcription factor nf-e2 subunit's dna2 binding domain from homo sapiens, northeast structural genomics3 consortium target hr4653b
17 d1svdm1
14.5
33
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
18 c2yyjA_
14.5
23
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxyphenylacetate-3-hydroxylase;PDBTitle: crystal structure of the oxygenase component (hpab) of 4-2 hydroxyphenylacetate 3-monooxygenase complexed with fad and 4-3 hydroxyphenylacetate
19 d2r40d1
14.3
47
Fold: Nuclear receptor ligand-binding domainSuperfamily: Nuclear receptor ligand-binding domainFamily: Nuclear receptor ligand-binding domain
20 d2csua3
14.1
24
Fold: Flavodoxin-likeSuperfamily: Succinyl-CoA synthetase domainsFamily: Succinyl-CoA synthetase domains
21 d1wdds_
not modelled
13.7
45
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
22 d1gk8i_
not modelled
13.6
40
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
23 c5mz2I_
not modelled
12.9
42
PDB header: photosynthesisChain: I: PDB Molecule: rubisco small subunit;PDBTitle: rubisco from thalassiosira antarctica
24 d1bwvs_
not modelled
12.4
58
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
25 c3oeoD_
not modelled
12.0
36
PDB header: signaling proteinChain: D: PDB Molecule: spheroplast protein y;PDBTitle: the crystal structure e. coli spy
26 c4a8xB_
not modelled
11.8
67
PDB header: transcriptionChain: B: PDB Molecule: hook-like, isoform a;PDBTitle: structure of the core asap complex
27 d1bxni_
not modelled
11.6
58
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
28 c6fkfd_
not modelled
11.5
12
PDB header: membrane proteinChain: D: PDB Molecule: atp synthase subunit beta, chloroplastic;PDBTitle: chloroplast f1fo conformation 1
29 c2ybvN_
not modelled
11.2
50
PDB header: lyaseChain: N: PDB Molecule: ribulose bisphosphate carboxylase small subunit;PDBTitle: structure of rubisco from thermosynechococcus elongatus
30 c5nv3P_
not modelled
11.0
50
PDB header: lyaseChain: P: PDB Molecule: ribulose bisphosphate carboxylase small chain 1;PDBTitle: structure of rubisco from rhodobacter sphaeroides in complex with cabp
31 d3eaua1
not modelled
10.7
20
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)
32 d8ruci_
not modelled
10.7
45
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
33 d1uzdc1
not modelled
10.5
40
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
34 d2v6ai1
not modelled
10.4
40
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
35 d1ej7s_
not modelled
10.2
40
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
36 c2yclA_
not modelled
10.1
27
PDB header: transferaseChain: A: PDB Molecule: carbon monoxide dehydrogenase corrinoid/iron-sulfurPDBTitle: complete structure of the corrinoid,iron-sulfur protein including2 the n-terminal domain with a 4fe-4s cluster
37 c3bt5A_
not modelled
10.1
32
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein duf305;PDBTitle: crystal structure of duf305 fragment from deinococcus radiodurans
38 d1ru0a_
not modelled
9.8
33
Fold: DCoH-likeSuperfamily: PCD-likeFamily: PCD-like
39 d1gyxa_
not modelled
9.7
31
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: 4-oxalocrotonate tautomerase-like
40 c3lutA_
not modelled
9.6
20
PDB header: membrane proteinChain: A: PDB Molecule: voltage-gated potassium channel subunit beta-2;PDBTitle: a structural model for the full-length shaker potassium channel kv1.2
41 c6rdlP_
not modelled
9.3
31
PDB header: proton transportChain: P: PDB Molecule: mitochondrial atp synthase subunit oscp;PDBTitle: cryo-em structure of polytomella f-atp synthase, rotary substate 1b,2 monomer-masked refinement
42 d1fi3a_
not modelled
8.8
35
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c
43 d1mhyb_
not modelled
8.8
29
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ribonucleotide reductase-like
44 c3epvB_
not modelled
8.6
19
PDB header: metal binding proteinChain: B: PDB Molecule: nickel and cobalt resistance protein cnrr;PDBTitle: x-ray structure of the metal-sensor cnrx in both the apo- and copper-2 bound forms
45 c3m20A_
not modelled
8.6
27
PDB header: isomeraseChain: A: PDB Molecule: 4-oxalocrotonate tautomerase, putative;PDBTitle: crystal structure of dmpi from archaeoglobus fulgidus determined to2 2.37 angstroms resolution
46 c4kmfA_
not modelled
8.5
27
PDB header: transferase/dnaChain: A: PDB Molecule: interferon-inducible and double-stranded-dependent eif-PDBTitle: crystal structure of zalpha domain from carassius auratus pkz in2 complex with z-dna
47 c4lb6B_
not modelled
8.5
45
PDB header: transferase/dnaChain: B: PDB Molecule: protein kinase containing z-dna binding domains;PDBTitle: crystal structure of pkz zalpha in complex with ds(cg)6 (tetragonal2 form)
48 d2apla1
not modelled
8.4
25
Fold: PG0816-likeSuperfamily: PG0816-likeFamily: PG0816-like
49 c2lz1A_
not modelled
8.2
41
PDB header: transcriptionChain: A: PDB Molecule: nuclear factor erythroid 2-related factor 2;PDBTitle: solution nmr structure of the dna-binding domain of human nf-e2-2 related factor 2, northeast structural genomics consortium (nesg)3 target hr3520o
50 c6b8ho_
not modelled
8.2
31
PDB header: membrane proteinChain: O: PDB Molecule: atp synthase subunit 5, mitochondrial;PDBTitle: mosaic model of yeast mitochondrial atp synthase monomer
51 c4uf6L_
not modelled
8.1
46
PDB header: hydrolaseChain: L: PDB Molecule: nuclear factor related to kappa-b-binding protein;PDBTitle: uch-l5 in complex with ubiquitin-propargyl bound to an activating2 fragment of ino80g
52 c1q2iA_
not modelled
8.1
44
PDB header: antitumor proteinChain: A: PDB Molecule: pnc27;PDBTitle: nmr solution structure of a peptide from the mdm-2 binding2 domain of the p53 protein that is selectively cytotoxic to3 cancer cells
53 c5jffD_
not modelled
7.9
56
PDB header: transferaseChain: D: PDB Molecule: uncharacterized protein yhfg;PDBTitle: e. coli ecfict mutant g55r in complex with ecfica
54 c4djeE_
not modelled
7.6
24
PDB header: transferase/vitamin-binding proteinChain: E: PDB Molecule: corrinoid/iron-sulfur protein large subunit;PDBTitle: crystal structure of folate-bound corrinoid iron-sulfur protein2 (cfesp) in complex with its methyltransferase (metr), co-crystallized3 with folate
55 c4xdnA_
not modelled
7.6
47
PDB header: cell cycleChain: A: PDB Molecule: mau2 chromatid cohesion factor homolog;PDBTitle: crystal structure of scc4 in complex with scc2n
56 d2bgxa2
not modelled
7.5
17
Fold: N-acetylmuramoyl-L-alanine amidase-likeSuperfamily: N-acetylmuramoyl-L-alanine amidase-likeFamily: N-acetylmuramoyl-L-alanine amidase-like
57 c4mqvD_
not modelled
7.4
55
PDB header: protein bindingChain: D: PDB Molecule: swi/snf-related matrix-associated actin-dependent regulatorPDBTitle: crystal complex of rpa32c and smarcal1 n-terminus
58 c4mqvB_
not modelled
7.4
55
PDB header: protein bindingChain: B: PDB Molecule: swi/snf-related matrix-associated actin-dependent regulatorPDBTitle: crystal complex of rpa32c and smarcal1 n-terminus
59 c5t4oL_
not modelled
7.2
19
PDB header: hydrolaseChain: L: PDB Molecule: atp synthase subunit delta;PDBTitle: autoinhibited e. coli atp synthase state 1
60 d1tm9a_
not modelled
7.0
64
Fold: Hypothetical protein MG354Superfamily: Hypothetical protein MG354Family: Hypothetical protein MG354
61 d1c75a_
not modelled
7.0
11
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c
62 d351ca_
not modelled
7.0
18
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c
63 c4u5pA_
not modelled
6.9
21
PDB header: isomeraseChain: A: PDB Molecule: rhcc;PDBTitle: crystal structure of native rhcc (yp_702633.1) from rhodococcus jostii2 rha1 at 1.78 angstrom
64 c2lr4A_
not modelled
6.9
50
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: spbc2 prophage-derived uncharacterized protein yola;PDBTitle: nmr structure of the protein np_390037.1 from bacillus subtilis
65 c3nzzA_
not modelled
6.8
36
PDB header: cell invasionChain: A: PDB Molecule: cell invasion protein sipd;PDBTitle: crystal structure of the salmonella type iii secretion system tip2 protein sipd
66 c3qzcA_
not modelled
6.4
19
PDB header: signaling proteinChain: A: PDB Molecule: periplasmic protein cpxp;PDBTitle: structure of the periplasmic stress response protein cpxp
67 c3bdnB_
not modelled
6.4
36
PDB header: transcription/dnaChain: B: PDB Molecule: lambda repressor;PDBTitle: crystal structure of the lambda repressor
68 c3itfA_
not modelled
6.3
17
PDB header: signaling proteinChain: A: PDB Molecule: periplasmic adaptor protein cpxp;PDBTitle: structural basis for the inhibitory function of the cpxp adaptor2 protein
69 c2qw6A_
not modelled
6.2
25
PDB header: hydrolaseChain: A: PDB Molecule: aaa atpase, central region;PDBTitle: crystal structure of the c-terminal domain of an aaa atpase from2 enterococcus faecium do
70 d2qw6a1
not modelled
6.2
25
Fold: post-AAA+ oligomerization domain-likeSuperfamily: post-AAA+ oligomerization domain-likeFamily: MgsA/YrvN C-terminal domain-like
71 c5jfzF_
not modelled
5.9
57
PDB header: transferaseChain: F: PDB Molecule: uncharacterized protein yhfg;PDBTitle: e. coli ecfict in complex with ecfica mutant e28g
72 c5jfzB_
not modelled
5.9
57
PDB header: transferaseChain: B: PDB Molecule: uncharacterized protein yhfg;PDBTitle: e. coli ecfict in complex with ecfica mutant e28g
73 c3c6vB_
not modelled
5.8
36
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: probable tautomerase/dehalogenase au4130;PDBTitle: crystal structure of au4130/apc7354, a probable enzyme from the2 thermophilic fungus aspergillus fumigatus
74 c2l69A_
not modelled
5.8
50
PDB header: de novo proteinChain: A: PDB Molecule: rossmann 2x3 fold protein;PDBTitle: solution nmr structure of de novo designed protein, p-loop ntpase2 fold, northeast structural genomics consortium target or28
75 d1euva_
not modelled
5.7
19
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Adenain-like
76 c4l8iA_
not modelled
5.7
19
PDB header: immune systemChain: A: PDB Molecule: rsv epitope scaffold ffl_005;PDBTitle: crystal structure of rsv epitope scaffold ffl_005
77 d1k78a1
not modelled
5.6
54
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain
78 c2rrnA_
not modelled
5.5
53
PDB header: protein transportChain: A: PDB Molecule: probable secdf protein-export membrane protein;PDBTitle: solution structure of secdf periplasmic domain p4
79 c4wlpB_
not modelled
5.5
46
PDB header: protein bindingChain: B: PDB Molecule: nuclear factor related to kappa-b-binding protein;PDBTitle: crystal structure of uch37-nfrkb inhibited deubiquitylating complex
80 c4bolA_
not modelled
5.4
26
PDB header: hydrolaseChain: A: PDB Molecule: ampdh2;PDBTitle: crystal structure of ampdh2 from pseudomonas aeruginosa in2 complex with pentapeptide
81 d1ynra1
not modelled
5.4
29
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c
82 d1k0ia2
not modelled
5.4
50
Fold: FAD-linked reductases, C-terminal domainSuperfamily: FAD-linked reductases, C-terminal domainFamily: PHBH-like
83 c2yjfM_
not modelled
5.3
31
PDB header: motor proteinChain: M: PDB Molecule: mkl/myocardin-like protein 1;PDBTitle: oligomeric assembly of actin bound to mrtf-a
84 d1t2sa_
not modelled
5.3
16
Fold: SH3-like barrelSuperfamily: PAZ domainFamily: PAZ domain
85 d1sknp_
not modelled
5.2
44
Fold: A DNA-binding domain in eukaryotic transcription factorsSuperfamily: A DNA-binding domain in eukaryotic transcription factorsFamily: A DNA-binding domain in eukaryotic transcription factors
86 c2kktA_
not modelled
5.2
75
PDB header: transcriptionChain: A: PDB Molecule: ataxin-7-like protein 3;PDBTitle: solution structure of the sca7 domain of human ataxin-7-l3 protein
87 c5u0pU_
not modelled
5.2
26
PDB header: transcriptionChain: U: PDB Molecule: mediator complex subunit 21;PDBTitle: cryo-em structure of the transcriptional mediator
88 c1yybA_
not modelled
5.1
75
PDB header: apoptosisChain: A: PDB Molecule: programmed cell death protein 5;PDBTitle: solution structure of 1-26 fragment of human programmed2 cell death 5 protein
89 c4clvB_
not modelled
5.1
19
PDB header: metal binding proteinChain: B: PDB Molecule: nickel-cobalt-cadmium resistance protein nccx;PDBTitle: crystal structure of dodecylphosphocholine-solubilized nccx2 from cupriavidus metallidurans 31a
90 d2cqea1
not modelled
5.1
30
Fold: CCCH zinc fingerSuperfamily: CCCH zinc fingerFamily: CCCH zinc finger
91 c6h5lB_
not modelled
5.0
29
PDB header: oxidoreductaseChain: B: PDB Molecule: conserved hypothetical cytochrome protein;PDBTitle: kuenenia stuttgartiensis reducing hao-like protein complex2 kustc0457/kustc0458
92 c5lqzU_
not modelled
5.0
25
PDB header: hydrolaseChain: U: PDB Molecule: atp synthase oscp subunit;PDBTitle: structure of f-atpase from pichia angusta, state1