| 1 |
|
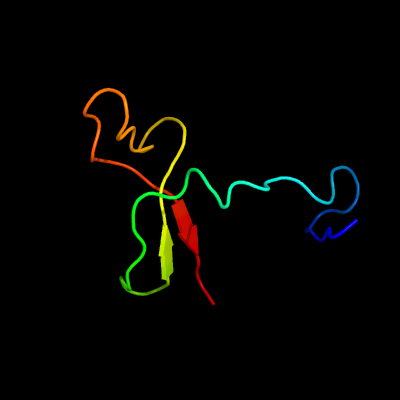
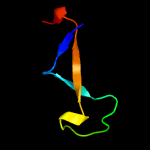
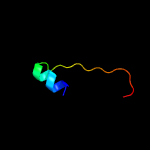
PDB 2dx5 chain A domain 1
Region: 32 - 85
Aligned: 44
Modelled: 54
Confidence: 53.4%
Identity: 20%
Fold: PH domain-like barrel
Superfamily: PH domain-like
Family: VPS36 N-terminal domain-like
Phyre2
| 2 |
|
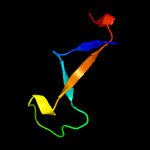
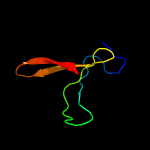
PDB 2mkc chain C
Region: 65 - 82
Aligned: 18
Modelled: 18
Confidence: 25.9%
Identity: 39%
PDB header:splicing
Chain: C: PDB Molecule:pre-mrna-splicing factor cwc26;
PDBTitle: cooperative structure of the heterotrimeric pre-mrna retention and2 splicing complex
Phyre2
| 3 |
|
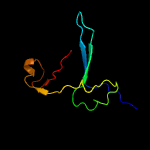
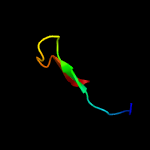
PDB 5zwo chain Y
Region: 65 - 82
Aligned: 18
Modelled: 18
Confidence: 24.4%
Identity: 39%
PDB header:splicing
Chain: Y: PDB Molecule:pre-mrna-splicing factor cwc26;
PDBTitle: cryo-em structure of the yeast b complex at average resolution of 3.92 angstrom
Phyre2
| 4 |
|
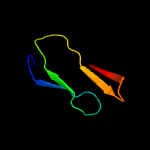
PDB 3ic8 chain D
Region: 42 - 85
Aligned: 44
Modelled: 44
Confidence: 22.1%
Identity: 25%
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:uncharacterized gst-like proteinprotein;
PDBTitle: the crystal structure of a gst-like protein from pseudomonas syringae2 to 2.4a
Phyre2
| 5 |
|
PDB 1yyp chain B
Region: 82 - 101
Aligned: 20
Modelled: 19
Confidence: 21.6%
Identity: 40%
PDB header:replication/transferase
Chain: B: PDB Molecule:dna polymerase;
PDBTitle: crystal structure of cytomegalovirus ul44 bound to c-terminal peptide2 from cmv ul54
Phyre2
| 6 |
|
PDB 1s21 chain A
Region: 80 - 112
Aligned: 33
Modelled: 33
Confidence: 19.5%
Identity: 27%
Fold: ADP-ribosylation
Superfamily: ADP-ribosylation
Family: AvrPphF ORF2, a type III effector
Phyre2
| 7 |
|
PDB 1s21 chain A
Region: 80 - 112
Aligned: 33
Modelled: 33
Confidence: 19.5%
Identity: 27%
PDB header:chaperone
Chain: A: PDB Molecule:orf2;
PDBTitle: crystal structure of avrpphf orf2, a type iii effector from p.2 syringae
Phyre2
| 8 |
|
PDB 5td3 chain B
Region: 14 - 88
Aligned: 74
Modelled: 75
Confidence: 18.0%
Identity: 14%
PDB header:oxidoreductase
Chain: B: PDB Molecule:catechol 1,2-dioxygenase;
PDBTitle: crystal structure of catechol 1,2-dioxygenase from burkholderia2 vietnamiensis
Phyre2
| 9 |
|
PDB 4a0t chain B
Region: 2 - 42
Aligned: 35
Modelled: 41
Confidence: 15.5%
Identity: 31%
PDB header:viral protein
Chain: B: PDB Molecule:tail fiber protein;
PDBTitle: structure of the carboxy-terminal domain of bacteriophage t7 fibre2 gp17 containing residues 371-553.
Phyre2
| 10 |
|
PDB 2orv chain B
Region: 32 - 67
Aligned: 35
Modelled: 36
Confidence: 13.6%
Identity: 20%
PDB header:transferase
Chain: B: PDB Molecule:thymidine kinase;
PDBTitle: human thymidine kinase 1 in complex with tp4a
Phyre2
| 11 |
|
PDB 3mp6 chain A
Region: 74 - 86
Aligned: 13
Modelled: 13
Confidence: 12.4%
Identity: 38%
PDB header:histone binding protein
Chain: A: PDB Molecule:maltose-binding periplasmic protein,linker,saga-associated
PDBTitle: complex structure of sgf29 and dimethylated h3k4
Phyre2
| 12 |
|
PDB 2bjq chain A domain 2
Region: 15 - 38
Aligned: 24
Modelled: 24
Confidence: 9.6%
Identity: 25%
Fold: MFPT repeat-like
Superfamily: MFPT repeat-like
Family: MFPT repeat
Phyre2
| 13 |
|
PDB 3icv chain A
Region: 86 - 116
Aligned: 29
Modelled: 31
Confidence: 8.7%
Identity: 14%
PDB header:hydrolase
Chain: A: PDB Molecule:lipase b;
PDBTitle: structural consequences of a circular permutation on lipase b from2 candida antartica
Phyre2
| 14 |
|
PDB 1t62 chain A
Region: 42 - 62
Aligned: 21
Modelled: 21
Confidence: 8.7%
Identity: 43%
Fold: PUA domain-like
Superfamily: PUA domain-like
Family: Hypothetical protein EF3133
Phyre2
| 15 |
|
PDB 2fno chain A domain 2
Region: 31 - 54
Aligned: 24
Modelled: 24
Confidence: 8.2%
Identity: 29%
Fold: Thioredoxin fold
Superfamily: Thioredoxin-like
Family: Glutathione S-transferase (GST), N-terminal domain
Phyre2
| 16 |
|
PDB 6j0f chain E
Region: 38 - 54
Aligned: 17
Modelled: 17
Confidence: 8.0%
Identity: 18%
PDB header:protein transport
Chain: E: PDB Molecule:pvc16;
PDBTitle: cryo-em structure of an extracellular contractile injection system,2 pvc sheath/tube terminator in extended state
Phyre2
| 17 |
|
PDB 6adq chain I
Region: 30 - 40
Aligned: 11
Modelled: 11
Confidence: 7.7%
Identity: 45%
PDB header:electron transport
Chain: I: PDB Molecule:cytochrome c oxidase subunit ctaj;
PDBTitle: respiratory complex ciii2civ2sod2 from mycobacterium smegmatis
Phyre2
| 18 |
|
PDB 1p5d chain X domain 1
Region: 47 - 69
Aligned: 23
Modelled: 23
Confidence: 6.7%
Identity: 17%
Fold: Phosphoglucomutase, first 3 domains
Superfamily: Phosphoglucomutase, first 3 domains
Family: Phosphoglucomutase, first 3 domains
Phyre2
| 19 |
|
PDB 4kf9 chain A
Region: 35 - 85
Aligned: 51
Modelled: 51
Confidence: 6.6%
Identity: 18%
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase protein;
PDBTitle: crystal structure of a glutathione transferase family member from2 ralstonia solanacearum, target efi-501780, with bound gsh coordinated3 to a zinc ion, ordered active site
Phyre2
| 20 |
|
PDB 3s9x chain A
Region: 42 - 62
Aligned: 21
Modelled: 20
Confidence: 6.4%
Identity: 24%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:asch domain;
PDBTitle: high resolution crystal structure of asch domain from lactobacillus2 crispatus jv v101
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|