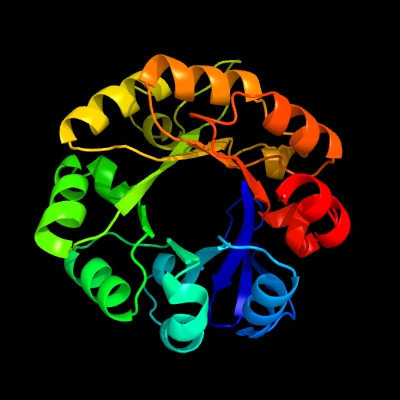
| 1 | d1qo2a_
|
|
 |
100.0 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
|
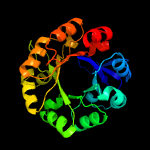
| 2 | c4axkB_
|
|
 |
100.0 |
62 |
PDB header:isomerase
Chain: B: PDB Molecule:1-(5-phosphoribosyl)-5-((5'-phosphoribosylamino)
PDBTitle: crystal structure of subhisa from the thermophile corynebacterium2 efficiens
|
|
|
|
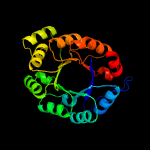
| 3 | d1jvna1
|
|
 |
100.0 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
|
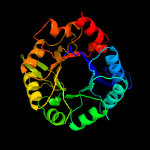
| 4 | c4x2rA_
|
|
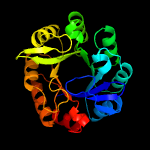 |
100.0 |
66 |
PDB header:isomerase
Chain: A: PDB Molecule:1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)
PDBTitle: crystal structure of pria from actinomyces urogenitalis
|
|
|
|
| 5 | c2y85D_
|
|
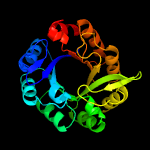 |
100.0 |
100 |
PDB header:isomerase
Chain: D: PDB Molecule:phosphoribosyl isomerase a;
PDBTitle: crystal structure of mycobacterium tuberculosis phosphoribosyl2 isomerase with bound rcdrp
|
|
|
|
| 6 | c1jvnB_
|
|
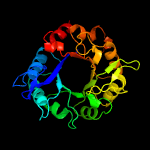 |
100.0 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:bifunctional histidine biosynthesis protein hishf;
PDBTitle: crystal structure of imidazole glycerol phosphate synthase: a tunnel2 through a (beta/alpha)8 barrel joins two active sites
|
|
|
|
| 7 | c4gj1A_
|
|
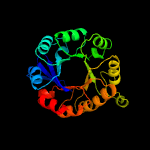 |
100.0 |
29 |
PDB header:isomerase
Chain: A: PDB Molecule:1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)
PDBTitle: crystal structure of 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)2 methylideneamino] imidazole-4-carboxamide isomerase (hisa).
|
|
|
|
| 8 | d1h5ya_
|
|
 |
100.0 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
|
| 9 | c4wd0A_
|
|
 |
100.0 |
62 |
PDB header:isomerase
Chain: A: PDB Molecule:1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)
PDBTitle: crystal structure of hisap form arthrobacter aurescens
|
|
|
|
| 10 | d1vzwa1
|
|
 |
100.0 |
69 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
|
| 11 | c3tdnB_
|
|
 |
100.0 |
23 |
PDB header:de novo protein
Chain: B: PDB Molecule:flr symmetric alpha-beta tim barrel;
PDBTitle: computationally designed two-fold symmetric tim-barrel protein, flr
|
|
|
|
| 12 | d1thfd_
|
|
 |
100.0 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
|
| 13 | c5aheA_
|
|
 |
100.0 |
32 |
PDB header:isomerase
Chain: A: PDB Molecule:
PDBTitle: crystal structure of salmonella enterica hisa
|
|
|
|
| 14 | d1ka9f_
|
|
 |
100.0 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
|
| 15 | c2agkA_
|
|
 |
100.0 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)
PDBTitle: structure of s. cerevisiae his6 protein
|
|
|
|
| 16 | c2w6rA_
|
|
 |
100.0 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:imidazole glycerol phosphate synthase subunit
PDBTitle: crystal structure of an artificial (ba)8-barrel protein2 designed from identical half barrels
|
|
|
|
| 17 | d1znna1
|
|
 |
100.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:PdxS-like |
|
|
|
| 18 | c1znnF_
|
|
 |
100.0 |
18 |
PDB header:biosynthetic protein
Chain: F: PDB Molecule:plp synthase;
PDBTitle: structure of the synthase subunit of plp synthase
|
|
|
|
| 19 | c4ml9A_
|
|
 |
100.0 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized tim barrel protein with the2 conserved phosphate binding site fromsebaldella termitidis
|
|
|
|
| 20 | c3tsmB_
|
|
 |
100.0 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of indole-3-glycerol phosphate synthase from2 brucella melitensis
|
|
|
|
| 21 | c3qjaA_ |
|
not modelled |
100.0 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of the mycobacterium tuberculosis indole-3-glycerol2 phosphate synthase (trpc) in apo form
|
|
|
| 22 | c6bmaA_ |
|
not modelled |
100.0 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: the crystal structure of indole-3-glycerol phosphate synthase from2 campylobacter jejuni subsp. jejuni nctc 11168
|
|
|
| 23 | d1a53a_ |
|
not modelled |
99.9 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 24 | d1vc4a_ |
|
not modelled |
99.9 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 25 | c2c3zA_ |
|
not modelled |
99.9 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of a truncated variant of indole-3-2 glycerol phosphate synthase from sulfolobus solfataricus
|
|
|
| 26 | c1piiA_ |
|
not modelled |
99.9 |
18 |
PDB header:bifunctional(isomerase and synthase)
Chain: A: PDB Molecule:n-(5'phosphoribosyl)anthranilate isomerase;
PDBTitle: three-dimensional structure of the bifunctional enzyme2 phosphoribosylanthranilate isomerase: indoleglycerolphosphate3 synthase from escherichia coli refined at 2.0 angstroms resolution
|
|
|
| 27 | d1i4na_ |
|
not modelled |
99.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 28 | d1j5ta_ |
|
not modelled |
99.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 29 | c3tdmD_ |
|
not modelled |
99.9 |
24 |
PDB header:de novo protein
Chain: D: PDB Molecule:computationally designed two-fold symmetric tim-barrel
PDBTitle: computationally designed tim-barrel protein, halfflr
|
|
|
| 30 | d1piia2 |
|
not modelled |
99.9 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 31 | c5umfB_ |
|
not modelled |
99.8 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:ribulose-phosphate 3-epimerase;
PDBTitle: crystal structure of a ribulose-phosphate 3-epimerase from neisseria2 gonorrhoeae with bound phosphate
|
|
|
| 32 | d2flia1 |
|
not modelled |
99.8 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 33 | c3q58A_ |
|
not modelled |
99.8 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structure of n-acetylmannosamine-6-phosphate epimerase from salmonella2 enterica
|
|
|
| 34 | d1tqja_ |
|
not modelled |
99.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 35 | d1h1ya_ |
|
not modelled |
99.8 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 36 | c3igsB_ |
|
not modelled |
99.8 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:n-acetylmannosamine-6-phosphate 2-epimerase 2;
PDBTitle: structure of the salmonella enterica n-acetylmannosamine-6-phosphate2 2-epimerase
|
|
|
| 37 | c5zjnB_ |
|
not modelled |
99.8 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:putative n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structure of n-acetylmannosamine-6-phosphate-2-epimerase from vibrio2 cholerae with n-acetylmannosamine-6-phosphate
|
|
|
| 38 | c4utwB_ |
|
not modelled |
99.8 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:putative n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structural characterisation of nane, mannac6p c2 epimerase,2 from clostridium perfingens
|
|
|
| 39 | d1y0ea_ |
|
not modelled |
99.7 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:NanE-like |
|
|
| 40 | d1yxya1 |
|
not modelled |
99.7 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:NanE-like |
|
|
| 41 | d1rpxa_ |
|
not modelled |
99.7 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 42 | c5zknA_ |
|
not modelled |
99.7 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:putative n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structure of n-acetylmannosamine-6-phosphate 2-epimerase from2 fusobacterium nucleatum
|
|
|
| 43 | c3inpA_ |
|
not modelled |
99.7 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:d-ribulose-phosphate 3-epimerase;
PDBTitle: 2.05 angstrom resolution crystal structure of d-ribulose-phosphate 3-2 epimerase from francisella tularensis.
|
|
|
| 44 | c3vkbA_ |
|
not modelled |
99.7 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:moeo5;
PDBTitle: crystal structure of moeo5 soaked with fspp overnight
|
|
|
| 45 | c6ei9A_ |
|
not modelled |
99.6 |
23 |
PDB header:flavoprotein
Chain: A: PDB Molecule:trna-dihydrouridine synthase b;
PDBTitle: crystal structure of e. coli trna-dihydrouridine synthase b (dusb)
|
|
|
| 46 | c5n2pA_ |
|
not modelled |
99.6 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: sulfolobus solfataricus tryptophan synthase a
|
|
|
| 47 | c5b69A_ |
|
not modelled |
99.6 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:geranylgeranylglyceryl phosphate synthase;
PDBTitle: crystal structure of geranylgeranylglyceryl phosphate synthase2 complexed with an g-1-p from thermoplasma acidophilum
|
|
|
| 48 | d1vhna_ |
|
not modelled |
99.6 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 49 | c3w9zA_ |
|
not modelled |
99.6 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:trna-dihydrouridine synthase c;
PDBTitle: crystal structure of dusc
|
|
|
| 50 | c4e38A_ |
|
not modelled |
99.6 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:keto-hydroxyglutarate-aldolase/keto-deoxy-phosphogluconate
PDBTitle: crystal structure of probable keto-hydroxyglutarate-aldolase from2 vibrionales bacterium swat-3 (target efi-502156)
|
|
|
| 51 | d1xm3a_ |
|
not modelled |
99.6 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:ThiG-like
Family:ThiG-like |
|
|
| 52 | c4xp7A_ |
|
not modelled |
99.5 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:trna-dihydrouridine(20) synthase [nad(p)+]-like;
PDBTitle: crystal structure of human trna dihydrouridine synthase 2
|
|
|
| 53 | c3cwoX_ |
|
not modelled |
99.5 |
20 |
PDB header:de novo protein
Chain: X: PDB Molecule:beta/alpha-barrel protein based on 1thf and 1tmy;
PDBTitle: a beta/alpha-barrel built by the combination of fragments from2 different folds
|
|
|
| 54 | d1wbha1 |
|
not modelled |
99.5 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 55 | c6oviA_ |
|
not modelled |
99.5 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:keto-deoxy-phosphogluconate aldolase;
PDBTitle: crystal structure of kdpg aldolase from legionella pneumophila with2 pyruvate captured at low ph as a covalent carbinolamine intermediate
|
|
|
| 56 | d1qopa_ |
|
not modelled |
99.5 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 57 | c4nu7C_ |
|
not modelled |
99.5 |
13 |
PDB header:isomerase
Chain: C: PDB Molecule:ribulose-phosphate 3-epimerase;
PDBTitle: 2.05 angstrom crystal structure of ribulose-phosphate 3-epimerase from2 toxoplasma gondii.
|
|
|
| 58 | c3b0vD_ |
|
not modelled |
99.5 |
24 |
PDB header:oxidoreductase/rna
Chain: D: PDB Molecule:trna-dihydrouridine synthase;
PDBTitle: trna-dihydrouridine synthase from thermus thermophilus in complex with2 trna
|
|
|
| 59 | d1rd5a_ |
|
not modelled |
99.5 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 60 | c4n6eA_ |
|
not modelled |
99.4 |
18 |
PDB header:lyase/biosynthetic protein
Chain: A: PDB Molecule:putative thiosugar synthase;
PDBTitle: crystal structure of amycolatopsis orientalis bexx/cyso complex
|
|
|
| 61 | d1vhca_ |
|
not modelled |
99.4 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 62 | c3qc3B_ |
|
not modelled |
99.4 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:d-ribulose-5-phosphate-3-epimerase;
PDBTitle: crystal structure of a d-ribulose-5-phosphate-3-epimerase (np_954699)2 from homo sapiens at 2.20 a resolution
|
|
|
| 63 | d1wa3a1 |
|
not modelled |
99.4 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 64 | c2v82A_ |
|
not modelled |
99.4 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxy-6-phosphogalactonate aldolase;
PDBTitle: kdpgal complexed to kdpgal
|
|
|
| 65 | d1tqxa_ |
|
not modelled |
99.4 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 66 | c5z9yB_ |
|
not modelled |
99.4 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:thiazole synthase;
PDBTitle: crystal structure of mycobacterium tuberculosis thiazole synthase2 (thig) complexed with dxp
|
|
|
| 67 | d1mxsa_ |
|
not modelled |
99.4 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 68 | c1zfjA_ |
|
not modelled |
99.4 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
|
|
|
| 69 | c3ct7E_ |
|
not modelled |
99.3 |
12 |
PDB header:isomerase
Chain: E: PDB Molecule:d-allulose-6-phosphate 3-epimerase;
PDBTitle: crystal structure of d-allulose 6-phosphate 3-epimerase2 from escherichia coli k-12
|
|
|
| 70 | c4bk9B_ |
|
not modelled |
99.3 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxo
PDBTitle: crystal structure of 2-keto-3-deoxy-6-phospho-gluconate aldolase from2 zymomonas mobilis atcc 29191
|
|
|
| 71 | d1xcfa_ |
|
not modelled |
99.3 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 72 | c4qccA_ |
|
not modelled |
99.3 |
17 |
PDB header:structural protein, lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxy-6-phosphogalactonate aldolase, peptidyl-
PDBTitle: structure of a cube-shaped, highly porous protein cage designed by2 fusing symmetric oligomeric domains
|
|
|
| 73 | c3labA_ |
|
not modelled |
99.3 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative kdpg (2-keto-3-deoxy-6-phosphogluconate) aldolase;
PDBTitle: crystal structure of a putative kdpg (2-keto-3-deoxy-6-2 phosphogluconate) aldolase from oleispira antarctica
|
|
|
| 74 | c3vndD_ |
|
not modelled |
99.2 |
18 |
PDB header:lyase
Chain: D: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha-subunit from the2 psychrophile shewanella frigidimarina k14-2
|
|
|
| 75 | c3navB_ |
|
not modelled |
99.2 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
|
|
|
| 76 | c5kzmA_ |
|
not modelled |
99.2 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha-beta chain complex from2 francisella tularensis
|
|
|
| 77 | c4fxsA_ |
|
not modelled |
99.2 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: inosine 5'-monophosphate dehydrogenase from vibrio cholerae complexed2 with imp and mycophenolic acid
|
|
|
| 78 | d1w0ma_ |
|
not modelled |
99.2 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 79 | d1geqa_ |
|
not modelled |
99.2 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 80 | c6nkeA_ |
|
not modelled |
99.2 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:geranylgeranylglyceryl phosphate synthase;
PDBTitle: wild-type gggps from thermoplasma volcanium
|
|
|
| 81 | d1xi3a_ |
|
not modelled |
99.2 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Thiamin phosphate synthase
Family:Thiamin phosphate synthase |
|
|
| 82 | c2ekcA_ |
|
not modelled |
99.2 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
|
|
|
| 83 | c4z87B_ |
|
not modelled |
99.2 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: structure of the imp dehydrogenase from ashbya gossypii bound to gdp
|
|
|
| 84 | c3thaB_ |
|
not modelled |
99.1 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: tryptophan synthase subunit alpha from campylobacter jejuni.
|
|
|
| 85 | d2tpsa_ |
|
not modelled |
99.1 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Thiamin phosphate synthase
Family:Thiamin phosphate synthase |
|
|
| 86 | c2z6jB_ |
|
not modelled |
99.1 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:trans-2-enoyl-acp reductase ii;
PDBTitle: crystal structure of s. pneumoniae enoyl-acyl carrier2 protein reductase (fabk) in complex with an inhibitor
|
|
|
| 87 | c5cssA_ |
|
not modelled |
99.1 |
8 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from thermoplasma2 acidophilum with glycerol 3-phosphate
|
|
|
| 88 | c5x8oA_ |
|
not modelled |
99.1 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: crystal structure of gmp reductase from trypanosoma brucei with2 guanosine 5'-triphosphate
|
|
|
| 89 | c3khjE_ |
|
not modelled |
99.1 |
17 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:inosine-5-monophosphate dehydrogenase;
PDBTitle: c. parvum inosine monophosphate dehydrogenase bound by inhibitor c64
|
|
|
| 90 | c2h6rG_ |
|
not modelled |
99.1 |
15 |
PDB header:isomerase
Chain: G: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase (tim) from2 methanocaldococcus jannaschii
|
|
|
| 91 | c3tsdA_ |
|
not modelled |
99.1 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: crystal structure of inosine-5'-monophosphate dehydrogenase from2 bacillus anthracis str. ames complexed with xmp
|
|
|
| 92 | d1hg3a_ |
|
not modelled |
99.1 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 93 | d1wv2a_ |
|
not modelled |
99.1 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:ThiG-like
Family:ThiG-like |
|
|
| 94 | c3f4wA_ |
|
not modelled |
99.1 |
19 |
PDB header:synthase, lyase
Chain: A: PDB Molecule:putative hexulose 6 phosphate synthase;
PDBTitle: the 1.65a crystal structure of 3-hexulose-6-phosphate2 synthase from salmonella typhimurium
|
|
|
| 95 | c3gr7A_ |
|
not modelled |
99.1 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dehydrogenase;
PDBTitle: structure of oye from geobacillus kaustophilus, hexagonal crystal form
|
|
|
| 96 | c3bo9B_ |
|
not modelled |
99.1 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative nitroalkan dioxygenase;
PDBTitle: crystal structure of putative nitroalkan dioxygenase (tm0800) from2 thermotoga maritima at 2.71 a resolution
|
|
|
| 97 | c3o63B_ |
|
not modelled |
99.1 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:probable thiamine-phosphate pyrophosphorylase;
PDBTitle: crystal structure of thiamin phosphate synthase from mycobacterium2 tuberculosis
|
|
|
| 98 | c2htmB_ |
|
not modelled |
99.0 |
19 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:thiazole biosynthesis protein thig;
PDBTitle: crystal structure of ttha0676 from thermus thermophilus hb8
|
|
|
| 99 | c3ajxA_ |
|
not modelled |
99.0 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:3-hexulose-6-phosphate synthase;
PDBTitle: crystal structure of 3-hexulose-6-phosphate synthase
|
|
|
| 100 | c4af0B_ |
|
not modelled |
99.0 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: crystal structure of cryptococcal inosine monophosphate2 dehydrogenase
|
|
|
| 101 | c3r2gA_ |
|
not modelled |
99.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine 5'-monophosphate dehydrogenase;
PDBTitle: crystal structure of inosine 5' monophosphate dehydrogenase from2 legionella pneumophila
|
|
|
| 102 | c4iqlB_ |
|
not modelled |
99.0 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:enoyl-(acyl-carrier-protein) reductase ii;
PDBTitle: crystal structure of porphyromonas gingivalis enoyl-acp reductase ii2 (fabk) with cofactors nadph and fmn
|
|
|
| 103 | c4dqwB_ |
|
not modelled |
99.0 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: crystal structure analysis of pa3770
|
|
|
| 104 | c4jejA_ |
|
not modelled |
99.0 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:geranylgeranylglyceryl phosphate synthase;
PDBTitle: gggps from flavobacterium johnsoniae
|
|
|
| 105 | c2rdtA_ |
|
not modelled |
99.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hydroxyacid oxidase 1;
PDBTitle: crystal structure of human glycolate oxidase (go) in complex with cdst
|
|
|
| 106 | c2gjlA_ |
|
not modelled |
99.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical protein pa1024;
PDBTitle: crystal structure of 2-nitropropane dioxygenase
|
|
|
| 107 | d1jcna1 |
|
not modelled |
99.0 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Inosine monophosphate dehydrogenase (IMPDH)
Family:Inosine monophosphate dehydrogenase (IMPDH) |
|
|
| 108 | c3ffsC_ |
|
not modelled |
99.0 |
29 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:inosine-5-monophosphate dehydrogenase;
PDBTitle: the crystal structure of cryptosporidium parvum inosine-5'-2 monophosphate dehydrogenase
|
|
|
| 109 | c5ey5A_ |
|
not modelled |
99.0 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:lbcats-a;
PDBTitle: lbcats
|
|
|
| 110 | c3cu2A_ |
|
not modelled |
99.0 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:ribulose-5-phosphate 3-epimerase;
PDBTitle: crystal structure of ribulose-5-phosphate 3-epimerase (yp_718263.1)2 from haemophilus somnus 129pt at 1.91 a resolution
|
|
|
| 111 | c4mm1E_ |
|
not modelled |
99.0 |
17 |
PDB header:transferase
Chain: E: PDB Molecule:geranylgeranylglyceryl phosphate synthase;
PDBTitle: gggps from methanothermobacter thermautotrophicus
|
|
|
| 112 | c4zqrD_ |
|
not modelled |
99.0 |
24 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:inosine-5'-monophosphate dehydrogenase,inosine-5'-
PDBTitle: crystal structure of the catalytic domain of the inosine monophosphate2 dehydrogenase from mycobacterium tuberculosis
|
|
|
| 113 | c2qr6A_ |
|
not modelled |
99.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:imp dehydrogenase/gmp reductase;
PDBTitle: crystal structure of imp dehydrogenase/gmp reductase-like protein2 (np_599840.1) from corynebacterium glutamicum atcc 13032 kitasato at3 1.50 a resolution
|
|
|
| 114 | d1pvna1 |
|
not modelled |
99.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Inosine monophosphate dehydrogenase (IMPDH)
Family:Inosine monophosphate dehydrogenase (IMPDH) |
|
|
| 115 | c4q33F_ |
|
not modelled |
98.9 |
18 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: crystal structure of inosine 5'-monophosphate dehydrogenase from2 clostridium perfringens complexed with imp and a110
|
|
|
| 116 | c4ff0B_ |
|
not modelled |
98.9 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: inosine 5'-monophosphate dehydrogenase from vibrio cholerae, deletion2 mutant, complexed with imp
|
|
|
| 117 | c5kinC_ |
|
not modelled |
98.9 |
16 |
PDB header:lyase
Chain: C: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha beta complex from2 streptococcus pneumoniae
|
|
|
| 118 | d1gtea2 |
|
not modelled |
98.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 119 | c2yw3E_ |
|
not modelled |
98.9 |
18 |
PDB header:lyase
Chain: E: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase/2-deydro-3-
PDBTitle: crystal structure analysis of the 4-hydroxy-2-oxoglutarate aldolase/2-2 deydro-3-deoxyphosphogluconate aldolase from tthb1
|
|
|
| 120 | d1zfja1 |
|
not modelled |
98.9 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Inosine monophosphate dehydrogenase (IMPDH)
Family:Inosine monophosphate dehydrogenase (IMPDH) |
|
|