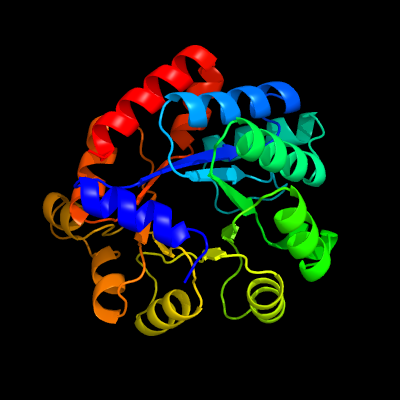
| 1 | c5tchG_
|
|
 |
100.0 |
98 |
PDB header:lyase
Chain: G: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase from m. tuberculosis -2 ligand-free form, trpa-g66v mutant
|
|
|
|
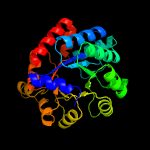
| 2 | c5k9xA_
|
|
 |
100.0 |
32 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha chain from legionella2 pneumophila subsp. pneumophila
|
|
|
|
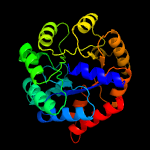
| 3 | c2ekcA_
|
|
 |
100.0 |
37 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
|
|
|
|
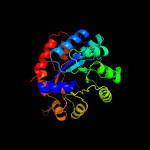
| 4 | c5kzmA_
|
|
 |
100.0 |
26 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha-beta chain complex from2 francisella tularensis
|
|
|
|
| 5 | c5ey5A_
|
|
 |
100.0 |
41 |
PDB header:lyase
Chain: A: PDB Molecule:lbcats-a;
PDBTitle: lbcats
|
|
|
|
| 6 | c3vndD_
|
|
 |
100.0 |
25 |
PDB header:lyase
Chain: D: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha-subunit from the2 psychrophile shewanella frigidimarina k14-2
|
|
|
|
| 7 | c3navB_
|
|
 |
100.0 |
29 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
|
|
|
|
| 8 | d1ujpa_
|
|
 |
100.0 |
44 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
|
| 9 | c5kinC_
|
|
 |
100.0 |
31 |
PDB header:lyase
Chain: C: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha beta complex from2 streptococcus pneumoniae
|
|
|
|
| 10 | d1qopa_
|
|
 |
100.0 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
|
| 11 | c3thaB_
|
|
 |
100.0 |
29 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: tryptophan synthase subunit alpha from campylobacter jejuni.
|
|
|
|
| 12 | d1geqa_
|
|
 |
100.0 |
31 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
|
| 13 | d1rd5a_
|
|
 |
100.0 |
33 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
|
| 14 | d1xcfa_
|
|
 |
100.0 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
|
| 15 | c5n2pA_
|
|
 |
100.0 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: sulfolobus solfataricus tryptophan synthase a
|
|
|
|
| 16 | c4jejA_
|
|
 |
100.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:geranylgeranylglyceryl phosphate synthase;
PDBTitle: gggps from flavobacterium johnsoniae
|
|
|
|
| 17 | d1viza_
|
|
 |
100.0 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
|
| 18 | d2f6ua1
|
|
 |
100.0 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
|
| 19 | c4naeA_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:heptaprenylglyceryl phosphate synthase;
PDBTitle: pcrb from geobacillus kaustophilus, with bound g1p
|
|
|
|
| 20 | c3w01A_
|
|
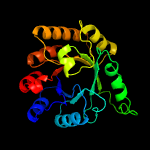 |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:heptaprenylglyceryl phosphate synthase;
PDBTitle: crystal structure of pcrb complexed with peg from staphylococcus2 aureus subsp. aureus mu3
|
|
|
|
| 21 | c3vkbA_ |
|
not modelled |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:moeo5;
PDBTitle: crystal structure of moeo5 soaked with fspp overnight
|
|
|
| 22 | c6nkeA_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:geranylgeranylglyceryl phosphate synthase;
PDBTitle: wild-type gggps from thermoplasma volcanium
|
|
|
| 23 | c5b69A_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:geranylgeranylglyceryl phosphate synthase;
PDBTitle: crystal structure of geranylgeranylglyceryl phosphate synthase2 complexed with an g-1-p from thermoplasma acidophilum
|
|
|
| 24 | d1j5ta_ |
|
not modelled |
100.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 25 | c3igsB_ |
|
not modelled |
100.0 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:n-acetylmannosamine-6-phosphate 2-epimerase 2;
PDBTitle: structure of the salmonella enterica n-acetylmannosamine-6-phosphate2 2-epimerase
|
|
|
| 26 | c3q58A_ |
|
not modelled |
100.0 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structure of n-acetylmannosamine-6-phosphate epimerase from salmonella2 enterica
|
|
|
| 27 | c2c3zA_ |
|
not modelled |
99.9 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of a truncated variant of indole-3-2 glycerol phosphate synthase from sulfolobus solfataricus
|
|
|
| 28 | c4nu7C_ |
|
not modelled |
99.9 |
11 |
PDB header:isomerase
Chain: C: PDB Molecule:ribulose-phosphate 3-epimerase;
PDBTitle: 2.05 angstrom crystal structure of ribulose-phosphate 3-epimerase from2 toxoplasma gondii.
|
|
|
| 29 | c5zjnB_ |
|
not modelled |
99.9 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:putative n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structure of n-acetylmannosamine-6-phosphate-2-epimerase from vibrio2 cholerae with n-acetylmannosamine-6-phosphate
|
|
|
| 30 | d1rpxa_ |
|
not modelled |
99.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 31 | d1tqja_ |
|
not modelled |
99.9 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 32 | c3qc3B_ |
|
not modelled |
99.9 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:d-ribulose-5-phosphate-3-epimerase;
PDBTitle: crystal structure of a d-ribulose-5-phosphate-3-epimerase (np_954699)2 from homo sapiens at 2.20 a resolution
|
|
|
| 33 | c3ct7E_ |
|
not modelled |
99.9 |
13 |
PDB header:isomerase
Chain: E: PDB Molecule:d-allulose-6-phosphate 3-epimerase;
PDBTitle: crystal structure of d-allulose 6-phosphate 3-epimerase2 from escherichia coli k-12
|
|
|
| 34 | c5zknA_ |
|
not modelled |
99.9 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:putative n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structure of n-acetylmannosamine-6-phosphate 2-epimerase from2 fusobacterium nucleatum
|
|
|
| 35 | d1y0ea_ |
|
not modelled |
99.9 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:NanE-like |
|
|
| 36 | d1h1ya_ |
|
not modelled |
99.9 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 37 | d1tqxa_ |
|
not modelled |
99.9 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 38 | d1q6oa_ |
|
not modelled |
99.9 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
|
|
| 39 | d1yxya1 |
|
not modelled |
99.9 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:NanE-like |
|
|
| 40 | c4utwB_ |
|
not modelled |
99.9 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:putative n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structural characterisation of nane, mannac6p c2 epimerase,2 from clostridium perfingens
|
|
|
| 41 | c3jr2D_ |
|
not modelled |
99.9 |
17 |
PDB header:biosynthetic protein
Chain: D: PDB Molecule:hexulose-6-phosphate synthase sgbh;
PDBTitle: x-ray crystal structure of the mg-bound 3-keto-l-gulonate-6-phosphate2 decarboxylase from vibrio cholerae o1 biovar el tor str. n16961
|
|
|
| 42 | d1piia2 |
|
not modelled |
99.9 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 43 | d1dvja_ |
|
not modelled |
99.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
|
|
| 44 | c4muzA_ |
|
not modelled |
99.9 |
17 |
PDB header:lyase/lyase inhibitor
Chain: A: PDB Molecule:orotidine 5'-phosphate decarboxylase;
PDBTitle: crystal structure of orotidine 5'-monophosphate decarboxylase from2 archaeoglobus fulgidus complexed with inhibitor bmp
|
|
|
| 45 | c3f4wA_ |
|
not modelled |
99.9 |
21 |
PDB header:synthase, lyase
Chain: A: PDB Molecule:putative hexulose 6 phosphate synthase;
PDBTitle: the 1.65a crystal structure of 3-hexulose-6-phosphate2 synthase from salmonella typhimurium
|
|
|
| 46 | c3inpA_ |
|
not modelled |
99.9 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:d-ribulose-phosphate 3-epimerase;
PDBTitle: 2.05 angstrom resolution crystal structure of d-ribulose-phosphate 3-2 epimerase from francisella tularensis.
|
|
|
| 47 | c3ajxA_ |
|
not modelled |
99.9 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:3-hexulose-6-phosphate synthase;
PDBTitle: crystal structure of 3-hexulose-6-phosphate synthase
|
|
|
| 48 | c5umfB_ |
|
not modelled |
99.9 |
19 |
PDB header:isomerase
Chain: B: PDB Molecule:ribulose-phosphate 3-epimerase;
PDBTitle: crystal structure of a ribulose-phosphate 3-epimerase from neisseria2 gonorrhoeae with bound phosphate
|
|
|
| 49 | c5z9yB_ |
|
not modelled |
99.8 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:thiazole synthase;
PDBTitle: crystal structure of mycobacterium tuberculosis thiazole synthase2 (thig) complexed with dxp
|
|
|
| 50 | d2czda1 |
|
not modelled |
99.8 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
|
|
| 51 | d2flia1 |
|
not modelled |
99.8 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 52 | c4luiB_ |
|
not modelled |
99.8 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:orotidine 5'-phosphate decarboxylase;
PDBTitle: crystal structure of orotidine 5'-monophosphate decarboxylase from2 methanocaldococcus jannaschii
|
|
|
| 53 | c3exsB_ |
|
not modelled |
99.8 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:rmpd (hexulose-6-phosphate synthase);
PDBTitle: crystal structure of kgpdc from streptococcus mutans in2 complex with d-r5p
|
|
|
| 54 | d1i4na_ |
|
not modelled |
99.7 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 55 | d1km4a_ |
|
not modelled |
99.7 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
|
|
| 56 | c4e38A_ |
|
not modelled |
99.7 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:keto-hydroxyglutarate-aldolase/keto-deoxy-phosphogluconate
PDBTitle: crystal structure of probable keto-hydroxyglutarate-aldolase from2 vibrionales bacterium swat-3 (target efi-502156)
|
|
|
| 57 | d1vc4a_ |
|
not modelled |
99.7 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 58 | d1vzwa1 |
|
not modelled |
99.7 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
| 59 | c4n6eA_ |
|
not modelled |
99.7 |
16 |
PDB header:lyase/biosynthetic protein
Chain: A: PDB Molecule:putative thiosugar synthase;
PDBTitle: crystal structure of amycolatopsis orientalis bexx/cyso complex
|
|
|
| 60 | c3qjaA_ |
|
not modelled |
99.7 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of the mycobacterium tuberculosis indole-3-glycerol2 phosphate synthase (trpc) in apo form
|
|
|
| 61 | d2tpsa_ |
|
not modelled |
99.6 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Thiamin phosphate synthase
Family:Thiamin phosphate synthase |
|
|
| 62 | c4ml9A_ |
|
not modelled |
99.6 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized tim barrel protein with the2 conserved phosphate binding site fromsebaldella termitidis
|
|
|
| 63 | c3tsmB_ |
|
not modelled |
99.5 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of indole-3-glycerol phosphate synthase from2 brucella melitensis
|
|
|
| 64 | d1thfd_ |
|
not modelled |
99.5 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
| 65 | d1wa3a1 |
|
not modelled |
99.5 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 66 | d1a53a_ |
|
not modelled |
99.5 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 67 | c6bmaA_ |
|
not modelled |
99.5 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: the crystal structure of indole-3-glycerol phosphate synthase from2 campylobacter jejuni subsp. jejuni nctc 11168
|
|
|
| 68 | d1w0ma_ |
|
not modelled |
99.5 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 69 | d1vhca_ |
|
not modelled |
99.4 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 70 | c4axkB_ |
|
not modelled |
99.4 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:1-(5-phosphoribosyl)-5-((5'-phosphoribosylamino)
PDBTitle: crystal structure of subhisa from the thermophile corynebacterium2 efficiens
|
|
|
| 71 | d1h5ya_ |
|
not modelled |
99.4 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
| 72 | c4gj1A_ |
|
not modelled |
99.4 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)
PDBTitle: crystal structure of 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)2 methylideneamino] imidazole-4-carboxamide isomerase (hisa).
|
|
|
| 73 | d1hg3a_ |
|
not modelled |
99.4 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 74 | d1wbha1 |
|
not modelled |
99.4 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 75 | c4x2rA_ |
|
not modelled |
99.3 |
23 |
PDB header:isomerase
Chain: A: PDB Molecule:1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)
PDBTitle: crystal structure of pria from actinomyces urogenitalis
|
|
|
| 76 | d1mxsa_ |
|
not modelled |
99.3 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 77 | c3ru6C_ |
|
not modelled |
99.3 |
12 |
PDB header:lyase
Chain: C: PDB Molecule:orotidine 5'-phosphate decarboxylase;
PDBTitle: 1.8 angstrom resolution crystal structure of orotidine 5'-phosphate2 decarboxylase (pyrf) from campylobacter jejuni subsp. jejuni nctc3 11168
|
|
|
| 78 | c5cssA_ |
|
not modelled |
99.3 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from thermoplasma2 acidophilum with glycerol 3-phosphate
|
|
|
| 79 | d1xi3a_ |
|
not modelled |
99.2 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Thiamin phosphate synthase
Family:Thiamin phosphate synthase |
|
|
| 80 | c4iqlB_ |
|
not modelled |
99.2 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:enoyl-(acyl-carrier-protein) reductase ii;
PDBTitle: crystal structure of porphyromonas gingivalis enoyl-acp reductase ii2 (fabk) with cofactors nadph and fmn
|
|
|
| 81 | c2v82A_ |
|
not modelled |
99.2 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxy-6-phosphogalactonate aldolase;
PDBTitle: kdpgal complexed to kdpgal
|
|
|
| 82 | c2y85D_ |
|
not modelled |
99.2 |
20 |
PDB header:isomerase
Chain: D: PDB Molecule:phosphoribosyl isomerase a;
PDBTitle: crystal structure of mycobacterium tuberculosis phosphoribosyl2 isomerase with bound rcdrp
|
|
|
| 83 | c3o63B_ |
|
not modelled |
99.2 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:probable thiamine-phosphate pyrophosphorylase;
PDBTitle: crystal structure of thiamin phosphate synthase from mycobacterium2 tuberculosis
|
|
|
| 84 | c6oviA_ |
|
not modelled |
99.2 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:keto-deoxy-phosphogluconate aldolase;
PDBTitle: crystal structure of kdpg aldolase from legionella pneumophila with2 pyruvate captured at low ph as a covalent carbinolamine intermediate
|
|
|
| 85 | d1ka9f_ |
|
not modelled |
99.2 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
| 86 | c5aheA_ |
|
not modelled |
99.2 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:
PDBTitle: crystal structure of salmonella enterica hisa
|
|
|
| 87 | c1piiA_ |
|
not modelled |
99.1 |
17 |
PDB header:bifunctional(isomerase and synthase)
Chain: A: PDB Molecule:n-(5'phosphoribosyl)anthranilate isomerase;
PDBTitle: three-dimensional structure of the bifunctional enzyme2 phosphoribosylanthranilate isomerase: indoleglycerolphosphate3 synthase from escherichia coli refined at 2.0 angstroms resolution
|
|
|
| 88 | c3tdnB_ |
|
not modelled |
99.1 |
11 |
PDB header:de novo protein
Chain: B: PDB Molecule:flr symmetric alpha-beta tim barrel;
PDBTitle: computationally designed two-fold symmetric tim-barrel protein, flr
|
|
|
| 89 | c4mm1E_ |
|
not modelled |
99.0 |
21 |
PDB header:transferase
Chain: E: PDB Molecule:geranylgeranylglyceryl phosphate synthase;
PDBTitle: gggps from methanothermobacter thermautotrophicus
|
|
|
| 90 | c2z6jB_ |
|
not modelled |
99.0 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:trans-2-enoyl-acp reductase ii;
PDBTitle: crystal structure of s. pneumoniae enoyl-acyl carrier2 protein reductase (fabk) in complex with an inhibitor
|
|
|
| 91 | c4b3yB_ |
|
not modelled |
99.0 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:fatty acid synthase;
PDBTitle: cryo-em structure of the mycobacterial fatty acid synthase
|
|
|
| 92 | c2cdh1_ |
|
not modelled |
98.9 |
20 |
PDB header:transferase
Chain: 1: PDB Molecule:enoyl reductase;
PDBTitle: architecture of the thermomyces lanuginosus fungal fatty2 acid synthase at 5 angstrom resolution.
|
|
|
| 93 | c4qccA_ |
|
not modelled |
98.9 |
21 |
PDB header:structural protein, lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxy-6-phosphogalactonate aldolase, peptidyl-
PDBTitle: structure of a cube-shaped, highly porous protein cage designed by2 fusing symmetric oligomeric domains
|
|
|
| 94 | c4bk9B_ |
|
not modelled |
98.9 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxo
PDBTitle: crystal structure of 2-keto-3-deoxy-6-phospho-gluconate aldolase from2 zymomonas mobilis atcc 29191
|
|
|
| 95 | c6b8sB_ |
|
not modelled |
98.9 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydroorotate dehydrogenase (quinone);
PDBTitle: crystal structure of dihydroorotate dehydrogenase from helicobacter2 pylori with bound fmn
|
|
|
| 96 | c4xq6A_ |
|
not modelled |
98.9 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydroorotate dehydrogenase (quinone);
PDBTitle: crystal structure of dihydroorotate dehydrogense from mycobacterium2 tuberculosis
|
|
|
| 97 | c2uvaI_ |
|
not modelled |
98.9 |
20 |
PDB header:transferase
Chain: I: PDB Molecule:fatty acid synthase beta subunits;
PDBTitle: crystal structure of fatty acid synthase from thermomyces2 lanuginosus at 3.1 angstrom resolution. this file contains3 the beta subunits of the fatty acid synthase. the entire4 crystal structure consists of one heterododecameric fatty5 acid synthase and is described in remark 400
|
|
|
| 98 | c2gjlA_ |
|
not modelled |
98.8 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical protein pa1024;
PDBTitle: crystal structure of 2-nitropropane dioxygenase
|
|
|
| 99 | c2rdtA_ |
|
not modelled |
98.8 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hydroxyacid oxidase 1;
PDBTitle: crystal structure of human glycolate oxidase (go) in complex with cdst
|
|
|
| 100 | c2htmB_ |
|
not modelled |
98.8 |
23 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:thiazole biosynthesis protein thig;
PDBTitle: crystal structure of ttha0676 from thermus thermophilus hb8
|
|
|
| 101 | c2vkzH_ |
|
not modelled |
98.8 |
17 |
PDB header:transferase
Chain: H: PDB Molecule:fatty acid synthase subunit beta;
PDBTitle: structure of the cerulenin-inhibited fungal fatty acid synthase type i2 multienzyme complex
|
|
|
| 102 | c4dbeB_ |
|
not modelled |
98.8 |
11 |
PDB header:lyase/lyase inhibitor
Chain: B: PDB Molecule:orotidine 5'-phosphate decarboxylase;
PDBTitle: crystal structure of orotidine 5'-monophosphate decarboxylase from2 sulfolobus solfataricus complexed with inhibitor bmp
|
|
|
| 103 | c3bo9B_ |
|
not modelled |
98.8 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative nitroalkan dioxygenase;
PDBTitle: crystal structure of putative nitroalkan dioxygenase (tm0800) from2 thermotoga maritima at 2.71 a resolution
|
|
|
| 104 | c4wd0A_ |
|
not modelled |
98.8 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)
PDBTitle: crystal structure of hisap form arthrobacter aurescens
|
|
|
| 105 | c1yadD_ |
|
not modelled |
98.8 |
16 |
PDB header:transcription
Chain: D: PDB Molecule:regulatory protein teni;
PDBTitle: structure of teni from bacillus subtilis
|
|
|
| 106 | c3labA_ |
|
not modelled |
98.8 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative kdpg (2-keto-3-deoxy-6-phosphogluconate) aldolase;
PDBTitle: crystal structure of a putative kdpg (2-keto-3-deoxy-6-2 phosphogluconate) aldolase from oleispira antarctica
|
|
|
| 107 | c1gthD_ |
|
not modelled |
98.8 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:dihydropyrimidine dehydrogenase;
PDBTitle: dihydropyrimidine dehydrogenase (dpd) from pig, ternary complex with2 nadph and 5-iodouracil
|
|
|
| 108 | c2h6rG_ |
|
not modelled |
98.7 |
18 |
PDB header:isomerase
Chain: G: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase (tim) from2 methanocaldococcus jannaschii
|
|
|
| 109 | d1goxa_ |
|
not modelled |
98.7 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 110 | c4zqrD_ |
|
not modelled |
98.7 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:inosine-5'-monophosphate dehydrogenase,inosine-5'-
PDBTitle: crystal structure of the catalytic domain of the inosine monophosphate2 dehydrogenase from mycobacterium tuberculosis
|
|
|
| 111 | d1tb3a1 |
|
not modelled |
98.7 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 112 | d1xm3a_ |
|
not modelled |
98.7 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:ThiG-like
Family:ThiG-like |
|
|
| 113 | d1juba_ |
|
not modelled |
98.7 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 114 | d1jcna1 |
|
not modelled |
98.7 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Inosine monophosphate dehydrogenase (IMPDH)
Family:Inosine monophosphate dehydrogenase (IMPDH) |
|
|
| 115 | c3tfxB_ |
|
not modelled |
98.7 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:orotidine 5'-phosphate decarboxylase;
PDBTitle: crystal structure of orotidine 5'-phosphate decarboxylase from2 lactobacillus acidophilus
|
|
|
| 116 | c5lsmF_ |
|
not modelled |
98.7 |
19 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:fmn-dependent nitronate monooxygenase;
PDBTitle: crystal structure of nitronate monooxygenase (so_0471) from shewanella2 oneidensis mr-1
|
|
|
| 117 | c3w9zA_ |
|
not modelled |
98.7 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:trna-dihydrouridine synthase c;
PDBTitle: crystal structure of dusc
|
|
|
| 118 | d1vrda1 |
|
not modelled |
98.7 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Inosine monophosphate dehydrogenase (IMPDH)
Family:Inosine monophosphate dehydrogenase (IMPDH) |
|
|
| 119 | c2a7nA_ |
|
not modelled |
98.7 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l(+)-mandelate dehydrogenase;
PDBTitle: crystal structure of the g81a mutant of the active chimera of (s)-2 mandelate dehydrogenase
|
|
|
| 120 | c3ivuB_ |
|
not modelled |
98.7 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:homocitrate synthase, mitochondrial;
PDBTitle: homocitrate synthase lys4 bound to 2-og
|
|
|