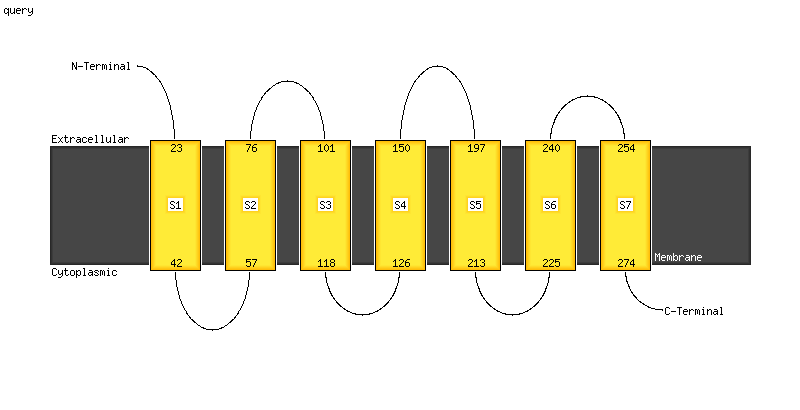
1 c5azcA_
100.0
22
PDB header: transferaseChain: A: PDB Molecule: prolipoprotein diacylglyceryl transferase;PDBTitle: crystal structure of escherichia coli lgt in complex with2 phosphatidylglycerol
2 c6m97A_
81.4
11
PDB header: transport proteinChain: A: PDB Molecule: chimera protein of high affinity copper uptake protein 1PDBTitle: crystal structure of the high-affinity copper transporter ctr1
3 c4kppA_
80.1
10
PDB header: membrane proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of h+/ca2+ exchanger cax
4 c5voxb_
62.8
11
PDB header: hydrolaseChain: B: PDB Molecule: v-type proton atpase subunit b;PDBTitle: yeast v-atpase in complex with legionella pneumophila effector sidk2 (rotational state 1)
5 c6dlnB_
62.4
20
PDB header: membrane proteinChain: B: PDB Molecule: transmembrane protein gp41;PDBTitle: oligomeric structure of the hiv gp41 mper-tmd in phospholipid bilayers
6 c6o7xa_
60.7
15
PDB header: membrane proteinChain: A: PDB Molecule: vacuolar atp synthase catalytic subunit a;PDBTitle: saccharomyces cerevisiae v-atpase stv1-v1vo state 3
7 c6qkcJ_
58.1
17
PDB header: membrane proteinChain: J: PDB Molecule: voltage-dependent calcium channel gamma-8 subunit;PDBTitle: glua1/2 in complex with auxiliary subunit gamma-8
8 c6cciA_
55.6
22
PDB header: transferaseChain: A: PDB Molecule: protein eskimo 1;PDBTitle: the crystal structure of xoat1
9 c1m57H_
50.9
20
PDB header: oxidoreductaseChain: H: PDB Molecule: cytochrome c oxidase;PDBTitle: structure of cytochrome c oxidase from rhodobacter2 sphaeroides (eq(i-286) mutant))
10 c5iwsA_
48.5
19
PDB header: transferaseChain: A: PDB Molecule: protein-n(pi)-phosphohistidine-sugar phosphotransferasePDBTitle: crystal structure of the transporter malt, the eiic domain from the2 maltose-specific phosphotransferase system
11 c5kzoA_
47.4
20
PDB header: transcriptionChain: A: PDB Molecule: neurogenic locus notch homolog protein 1;PDBTitle: notch1 transmembrane and associated juxtamembrane segment
12 c3m7eA_
46.4
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: tellurite resistance protein teha homolog;PDBTitle: crystal structure of plant slac1 homolog teha
13 c3m7bA_
46.4
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: tellurite resistance protein teha homolog;PDBTitle: crystal structure of plant slac1 homolog teha
14 c5a1sB_
44.4
15
PDB header: transport proteinChain: B: PDB Molecule: citrate-sodium symporter;PDBTitle: crystal structure of the sodium-dependent citrate symporter secits2 form salmonella enterica.
15 c6nbxG_
35.6
18
PDB header: oxidoreductaseChain: G: PDB Molecule: nadh-quinone oxidoreductase subunit j;PDBTitle: t.elongatus ndh (data-set 2)
16 c2k21A_
31.3
38
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily ePDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
17 c3nd0A_
29.8
14
PDB header: transport proteinChain: A: PDB Molecule: sll0855 protein;PDBTitle: x-ray crystal structure of a slow cyanobacterial cl-/h+ antiporter
18 c6e8wC_
28.2
20
PDB header: viral proteinChain: C: PDB Molecule: envelope glycoprotein gp160;PDBTitle: mper-tm domain of hiv-1 envelope glycoprotein (env)
19 c2m0qA_
27.4
10
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily e member 2;PDBTitle: solution nmr analysis of intact kcne2 in detergent micelles2 demonstrate a straight transmembrane helix
20 c6btmC_
27.1
16
PDB header: membrane proteinChain: C: PDB Molecule: alternative complex iii subunit c;PDBTitle: structure of alternative complex iii from flavobacterium johnsoniae2 (wild type)
21 c3wajA_
not modelled
25.7
12
PDB header: transferaseChain: A: PDB Molecule: transmembrane oligosaccharyl transferase;PDBTitle: crystal structure of the archaeoglobus fulgidus2 oligosaccharyltransferase (o29867_arcfu) complex with zn and sulfate
22 c2mg1A_
not modelled
25.0
22
PDB header: viral proteinChain: A: PDB Molecule: transmembrane protein gp41;PDBTitle: nmr assignment and structure of a peptide derived from the trans-2 membrane region of hiv-1 gp41 in the presence of3 hexafluoroisopropanol
23 c2ndjA_
not modelled
24.4
5
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily e member 3;PDBTitle: structural basis for kcne3 and estrogen modulation of the kcnq12 channel
24 c5t4oJ_
not modelled
23.5
9
PDB header: hydrolaseChain: J: PDB Molecule: atp synthase subunit b;PDBTitle: autoinhibited e. coli atp synthase state 1
25 c5yq7L_
not modelled
22.9
13
PDB header: photosynthesisChain: L: PDB Molecule: precursor for l subunits of photosynthetic reaction center;PDBTitle: cryo-em structure of the rc-lh core complex from roseiflexus2 castenholzii
26 c6f0kF_
not modelled
22.8
10
PDB header: membrane proteinChain: F: PDB Molecule: actf;PDBTitle: alternative complex iii
27 d2i5nl1
not modelled
22.6
20
Fold: Bacterial photosystem II reaction centre, L and M subunitsSuperfamily: Bacterial photosystem II reaction centre, L and M subunitsFamily: Bacterial photosystem II reaction centre, L and M subunits
28 c2miiA_
not modelled
22.4
13
PDB header: protein bindingChain: A: PDB Molecule: penicillin-binding protein activator lpob;PDBTitle: nmr structure of e. coli lpob
29 c2b6pA_
not modelled
21.5
13
PDB header: membrane proteinChain: A: PDB Molecule: lens fiber major intrinsic protein;PDBTitle: x-ray structure of lens aquaporin-0 (aqp0) (lens mip) in an open pore2 state
30 c3klzE_
not modelled
21.3
10
PDB header: membrane proteinChain: E: PDB Molecule: putative formate transporter 1;PDBTitle: pentameric formate channel with formate bound
31 c5ir6A_
not modelled
21.1
13
PDB header: oxidoreductaseChain: A: PDB Molecule: bd-type quinol oxidase subunit i;PDBTitle: the structure of bd oxidase from geobacillus thermodenitrificans
32 c4dveA_
not modelled
20.9
9
PDB header: transport proteinChain: A: PDB Molecule: biotin transporter bioy;PDBTitle: crystal structure at 2.1 a of the s-component for biotin from an ecf-2 type abc transporter
33 c2ls4A_
not modelled
20.8
11
PDB header: metal transportChain: A: PDB Molecule: high affinity copper uptake protein 1;PDBTitle: 1h chemical shift assignments for the third transmembrane domain from2 the human copper transport 1
34 c4o9uB_
not modelled
20.4
9
PDB header: membrane proteinChain: B: PDB Molecule: nad(p) transhydrogenase subunit beta;PDBTitle: mechanism of transhydrogenase coupling proton translocation and2 hydride transfer
35 d2j8cl1
not modelled
20.1
15
Fold: Bacterial photosystem II reaction centre, L and M subunitsSuperfamily: Bacterial photosystem II reaction centre, L and M subunitsFamily: Bacterial photosystem II reaction centre, L and M subunits
36 c5wsnC_
not modelled
19.5
19
PDB header: virusChain: C: PDB Molecule: e protein;PDBTitle: structure of japanese encephalitis virus
37 c2ht2B_
not modelled
18.9
11
PDB header: membrane proteinChain: B: PDB Molecule: h(+)/cl(-) exchange transporter clca;PDBTitle: structure of the escherichia coli clc chloride channel2 y445h mutant and fab complex
38 c6fkip_
not modelled
18.5
14
PDB header: membrane proteinChain: P: PDB Molecule: atp synthase subunit c, chloroplastic;PDBTitle: chloroplast f1fo conformation 3
39 c1m06J_
not modelled
18.4
43
PDB header: virus/dnaChain: J: PDB Molecule: small core protein;PDBTitle: structural studies of bacteriophage alpha3 assembly, x-ray2 crystallography
40 c1rb8J_
not modelled
18.3
43
PDB header: virus/dnaChain: J: PDB Molecule: small core protein;PDBTitle: the phix174 dna binding protein j in two different capsid2 environments.
41 c5xu1M_
not modelled
17.8
17
PDB header: transport proteinChain: M: PDB Molecule: abc transporter permeae;PDBTitle: structure of a non-canonical abc transporter from streptococcus2 pneumoniae r6
42 c2m59B_
not modelled
17.1
17
PDB header: transferaseChain: B: PDB Molecule: vascular endothelial growth factor receptor 2;PDBTitle: spatial structure of dimeric vegfr2 membrane domain in dpc micelles
43 c2m59A_
not modelled
17.1
17
PDB header: transferaseChain: A: PDB Molecule: vascular endothelial growth factor receptor 2;PDBTitle: spatial structure of dimeric vegfr2 membrane domain in dpc micelles
44 c4hzuS_
not modelled
16.9
15
PDB header: hydrolase, transport proteinChain: S: PDB Molecule: predicted membrane protein;PDBTitle: structure of a bacterial energy-coupling factor transporter
45 c4hydA_
not modelled
16.9
18
PDB header: membrane proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: structure of a presenilin family intramembrane aspartate protease in2 c2221 space group
46 d1kpla_
not modelled
16.9
14
Fold: Clc chloride channelSuperfamily: Clc chloride channelFamily: Clc chloride channel
47 c6bbjC_
not modelled
16.8
11
PDB header: metal transportChain: C: PDB Molecule: transient receptor potential cation channel, subfamily v,PDBTitle: xenopus tropicalis trpv4
48 c2yiuE_
not modelled
16.5
17
PDB header: oxidoreductaseChain: E: PDB Molecule: cytochrome c1, heme protein;PDBTitle: x-ray structure of the dimeric cytochrome bc1 complex from2 the soil bacterium paracoccus denitrificans at 2.73 angstrom resolution
49 c3kcvG_
not modelled
16.2
12
PDB header: transport proteinChain: G: PDB Molecule: probable formate transporter 1;PDBTitle: structure of formate channel
50 d1czan3
not modelled
16.1
43
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase
51 c5nikK_
not modelled
16.0
19
PDB header: transport proteinChain: K: PDB Molecule: macrolide export atp-binding/permease protein macb;PDBTitle: structure of the macab-tolc abc-type tripartite multidrug efflux pump
52 d1j4na_
not modelled
15.8
14
Fold: Aquaporin-likeSuperfamily: Aquaporin-likeFamily: Aquaporin-like
53 c6cfwF_
not modelled
15.7
10
PDB header: membrane proteinChain: F: PDB Molecule: monovalent cation/h+ antiporter subunit b;PDBTitle: cryoem structure of a respiratory membrane-bound hydrogenase
54 c5aexB_
not modelled
15.5
15
PDB header: membrane proteinChain: B: PDB Molecule: ammonium transporter mep2;PDBTitle: crystal structure of saccharomyces cerevisiae mep2
55 c2bpa3_
not modelled
15.5
43
PDB header: virus/dnaChain: 3: PDB Molecule: protein (subunit of bacteriophage phix174);PDBTitle: atomic structure of single-stranded dna bacteriophage2 phix174 and its functional implications
56 d1eysl_
not modelled
14.8
17
Fold: Bacterial photosystem II reaction centre, L and M subunitsSuperfamily: Bacterial photosystem II reaction centre, L and M subunitsFamily: Bacterial photosystem II reaction centre, L and M subunits
57 c1w9nA_
not modelled
14.6
11
PDB header: antibioticChain: A: PDB Molecule: epilancin 15x;PDBTitle: isolation and characterization of epilancin 15x, a novel antibiotic2 from a clinical strain of staphylococcus epidermidis
58 c6o7ua_
not modelled
14.6
11
PDB header: membrane proteinChain: A: PDB Molecule: PDBTitle: saccharomyces cerevisiae v-atpase stv1-vo
59 c5levA_
not modelled
14.4
15
PDB header: transferaseChain: A: PDB Molecule: udp-n-acetylglucosamine--dolichyl-phosphate n-PDBTitle: crystal structure of human udp-n-acetylglucosamine-dolichyl-phosphate2 n-acetylglucosaminephosphotransferase (dpagt1) (v264g mutant)
60 d1k8ke_
not modelled
14.3
9
Fold: Arp2/3 complex 21 kDa subunit ARPC3Superfamily: Arp2/3 complex 21 kDa subunit ARPC3Family: Arp2/3 complex 21 kDa subunit ARPC3
61 d1bg3a1
not modelled
14.1
43
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase
62 d1czan1
not modelled
14.1
43
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase
63 c5d7tC_
not modelled
14.1
10
PDB header: transport proteinChain: C: PDB Molecule: s-component for folate;PDBTitle: folate ecf transporter: apo state
64 c1p58C_
not modelled
13.6
18
PDB header: virusChain: C: PDB Molecule: major envelope protein e;PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
65 d1bg3a3
not modelled
13.5
50
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase
66 d1v4sa1
not modelled
13.5
50
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase
67 c2fynH_
not modelled
13.5
14
PDB header: oxidoreductaseChain: H: PDB Molecule: cytochrome c1;PDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
68 c2j3tB_
not modelled
13.4
8
PDB header: transportChain: B: PDB Molecule: trafficking protein particle complex subunit 6a;PDBTitle: the crystal structure of the bet3-trs33-bet5-trs23 complex.
69 c5aezA_
not modelled
13.1
13
PDB header: membrane proteinChain: A: PDB Molecule: mep2;PDBTitle: crystal structure of candida albicans mep2
70 c5nf8A_
not modelled
12.9
5
PDB header: membrane proteinChain: A: PDB Molecule: respiratory supercomplex factor 1, mitochondrial;PDBTitle: solution structure of detergent-solubilized rcf1, a yeast2 mitochondrial inner membrane protein involved in respiratory complex3 iii/iv supercomplex formation
71 c6nr3A_
not modelled
12.8
18
PDB header: transport proteinChain: A: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of the trpm8 ion channel in complex with high2 occupancy icilin, pi(4,5)p2, and calcium
72 c6nr3D_
not modelled
12.8
18
PDB header: transport proteinChain: D: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of the trpm8 ion channel in complex with high2 occupancy icilin, pi(4,5)p2, and calcium
73 c6nr3B_
not modelled
12.8
18
PDB header: transport proteinChain: B: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of the trpm8 ion channel in complex with high2 occupancy icilin, pi(4,5)p2, and calcium
74 c6nr3C_
not modelled
12.8
18
PDB header: transport proteinChain: C: PDB Molecule: transient receptor potential cation channel subfamily mPDBTitle: cryo-em structure of the trpm8 ion channel in complex with high2 occupancy icilin, pi(4,5)p2, and calcium
75 c4djiA_
not modelled
12.6
7
PDB header: transport proteinChain: A: PDB Molecule: probable glutamate/gamma-aminobutyrate antiporter;PDBTitle: structure of glutamate-gaba antiporter gadc
76 c6c5wA_
not modelled
12.3
15
PDB header: membrane proteinChain: A: PDB Molecule: calcium uniporter;PDBTitle: crystal structure of the mitochondrial calcium uniporter
77 c3tdsC_
not modelled
12.3
14
PDB header: membrane proteinChain: C: PDB Molecule: formate/nitrite transporter;PDBTitle: crystal structure of hsc f194i
78 c6btmD_
not modelled
12.2
13
PDB header: membrane proteinChain: D: PDB Molecule: alternative complex iii subunit d;PDBTitle: structure of alternative complex iii from flavobacterium johnsoniae2 (wild type)
79 c1zrtD_
not modelled
12.2
4
PDB header: oxidoreductase/metal transportChain: D: PDB Molecule: cytochrome c1;PDBTitle: rhodobacter capsulatus cytochrome bc1 complex with2 stigmatellin bound
80 d1eysm_
not modelled
12.1
17
Fold: Bacterial photosystem II reaction centre, L and M subunitsSuperfamily: Bacterial photosystem II reaction centre, L and M subunitsFamily: Bacterial photosystem II reaction centre, L and M subunits
81 c6dnfA_
not modelled
12.1
18
PDB header: membrane proteinChain: A: PDB Molecule: mitochondrial calcium uniporter mcu;PDBTitle: cryo-em structure of the mitochondrial calcium uniporter mcu from the2 fungus cyphellophora europaea
82 c6gcsJ_
not modelled
12.0
10
PDB header: oxidoreductaseChain: J: PDB Molecule: nujm subunit;PDBTitle: cryo-em structure of respiratory complex i from yarrowia lipolytica
83 c4b03A_
not modelled
11.8
13
PDB header: virusChain: A: PDB Molecule: dengue virus 1 e protein;PDBTitle: 6a electron cryomicroscopy structure of immature dengue virus serotype2 1
84 c6agfB_
not modelled
11.8
13
PDB header: membrane proteinChain: B: PDB Molecule: sodium channel subunit beta-1;PDBTitle: structure of the human voltage-gated sodium channel nav1.4 in complex2 with beta1
85 c5yq72_
not modelled
11.7
20
PDB header: photosynthesisChain: 2: PDB Molecule: beta subunit of light-harvesting 1;PDBTitle: cryo-em structure of the rc-lh core complex from roseiflexus2 castenholzii
86 c2jpjA_
not modelled
11.6
33
PDB header: antimicrobial proteinChain: A: PDB Molecule: bacteriocin lactococcin-g subunit alpha;PDBTitle: lactococcin g-a in dpc
87 c2mj2A_
not modelled
11.6
6
PDB header: viral proteinChain: A: PDB Molecule: agnoprotein;PDBTitle: structure of the dimerization domain of the human polyoma, jc virus2 agnoprotein is an amphipathic alpha-helix.
88 c3tijA_
not modelled
11.5
17
PDB header: membrane proteinChain: A: PDB Molecule: nupc family protein;PDBTitle: crystal structure of a concentrative nucleoside transporter from2 vibrio cholerae
89 c5zbgC_
not modelled
11.5
17
PDB header: membrane proteinChain: C: PDB Molecule: short transient receptor potential channel 3;PDBTitle: cryo-em structure of human trpc3 at 4.36a resolution
90 c6j5ib_
not modelled
11.4
10
PDB header: membrane proteinChain: B: PDB Molecule: atp synthase subunit alpha, mitochondrial;PDBTitle: cryo-em structure of the mammalian dp-state atp synthase
91 c6btmF_
not modelled
11.4
9
PDB header: membrane proteinChain: F: PDB Molecule: alternative complex iii subunit f;PDBTitle: structure of alternative complex iii from flavobacterium johnsoniae2 (wild type)
92 c3orgB_
not modelled
11.1
12
PDB header: transport proteinChain: B: PDB Molecule: cmclc;PDBTitle: crystal structure of a eukaryotic clc transporter
93 c1p84D_
not modelled
11.0
10
PDB header: oxidoreductaseChain: D: PDB Molecule: cytochrome c1, heme protein;PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
94 c3iyzA_
not modelled
10.9
18
PDB header: transport proteinChain: A: PDB Molecule: aquaporin-4;PDBTitle: structure of aquaporin-4 s180d mutant at 10.0 a resolution from2 electron micrograph
95 c5lnko_
not modelled
10.9
17
PDB header: oxidoreductaseChain: O: PDB Molecule: PDBTitle: entire ovine respiratory complex i
96 c5dirD_
not modelled
10.8
22
PDB header: hydrolaseChain: D: PDB Molecule: lipoprotein signal peptidase;PDBTitle: membrane protein at 2.8 angstroms
97 c5l2bC_
not modelled
10.8
16
PDB header: transport proteinChain: C: PDB Molecule: nucleoside permease;PDBTitle: structure of cntnw n149s, e332a in an outward-facing state
98 c3llqB_
not modelled
10.7
11
PDB header: membrane proteinChain: B: PDB Molecule: aquaporin z 2;PDBTitle: aquaporin structure from plant pathogen agrobacterium tumerfaciens
99 c3j8eG_
not modelled
10.7
9
PDB header: transport protein/isomeraseChain: G: PDB Molecule: ryanodine receptor 1;PDBTitle: cryo-em structure of ryanodine receptor/calstabin-2 complex