1 c6o7ua_
91.9
21
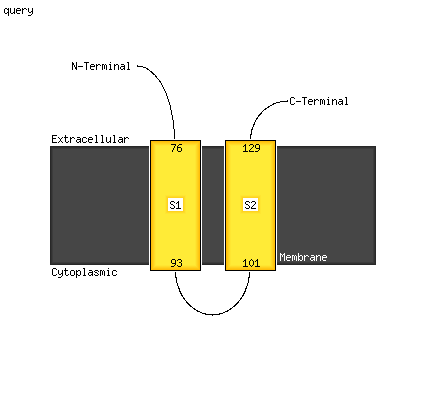
PDB header: membrane proteinChain: A: PDB Molecule: PDBTitle: saccharomyces cerevisiae v-atpase stv1-vo
2 c5gasN_
66.7
20
PDB header: hydrolaseChain: N: PDB Molecule: archaeal/vacuolar-type h+-atpase subunit i;PDBTitle: thermus thermophilus v/a-atpase, conformation 2
3 c5i1mV_
54.4
17
PDB header: membrane proteinChain: V: PDB Molecule: v-type proton atpase subunit a, vacuolar isoform;PDBTitle: yeast v-atpase average of densities, a subunit segment
4 d1kf6d_
52.4
25
Fold: Heme-binding four-helical bundleSuperfamily: Fumarate reductase respiratory complex transmembrane subunitsFamily: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)
5 c6o7xa_
43.4
28
PDB header: membrane proteinChain: A: PDB Molecule: vacuolar atp synthase catalytic subunit a;PDBTitle: saccharomyces cerevisiae v-atpase stv1-v1vo state 3
6 c5voxb_
25.7
16
PDB header: hydrolaseChain: B: PDB Molecule: v-type proton atpase subunit b;PDBTitle: yeast v-atpase in complex with legionella pneumophila effector sidk2 (rotational state 1)
7 c2aklA_
25.5
42
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: phna-like protein pa0128;PDBTitle: solution structure for phn-a like protein pa0128 from2 pseudomonas aeruginosa
8 d1epwa2
24.9
33
Fold: beta-TrefoilSuperfamily: STI-likeFamily: Clostridium neurotoxins, C-terminal domain
9 c1erfA_
24.0
42
PDB header: viral proteinChain: A: PDB Molecule: transmembrane glycoprotein;PDBTitle: conformational mapping of the n-terminal fusion peptide of2 hiv-1 gp41 using 13c-enhanced fourier transform infrared3 spectroscopy (ftir)
10 c2pjvA_
17.9
42
PDB header: viral proteinChain: A: PDB Molecule: envelope glycoprotein;PDBTitle: solution structure of hiv-1 gp41 fusion domain bound to dpc micelle
11 c5xvjB_
15.6
21
PDB header: gene regulationChain: B: PDB Molecule: phd finger protein alfin-like 7;PDBTitle: crystal structure of al7 pal domain
12 c5tj5A_
15.5
18
PDB header: motor proteinChain: A: PDB Molecule: v-type proton atpase subunit a;PDBTitle: atomic model for the membrane-embedded motor of a eukaryotic v-atpase
13 c5nwuA_
14.8
44
PDB header: viral proteinChain: A: PDB Molecule: wtfp-tag,gp41;PDBTitle: nmr assignment and structure of a peptide derived from the fusion2 peptide of hiv-1 gp41 in the presence of hexafluoroisopropanol
14 c6cfwB_
13.7
27
PDB header: membrane proteinChain: B: PDB Molecule: monovalent cation/h+ antiporter subunit f;PDBTitle: cryoem structure of a respiratory membrane-bound hydrogenase
15 c2jxfA_
11.9
23
PDB header: viral protein, membrane proteinChain: A: PDB Molecule: genome polyprotein;PDBTitle: the solution structure of hcv ns4b(40-69)
16 c2ariA_
8.8
22
PDB header: viral proteinChain: A: PDB Molecule: envelope polyprotein gp160;PDBTitle: solution structure of micelle-bound fusion domain of hiv-12 gp41
17 c4nl6C_
8.0
12
PDB header: splicingChain: C: PDB Molecule: survival motor neuron protein;PDBTitle: structure of the full-length form of the protein smn found in healthy2 patients
18 c1p5aA_
7.5
42
PDB header: viral proteinChain: A: PDB Molecule: envelope polyprotein gp160;PDBTitle: conformational mapping of the n-terminal peptide of hiv-12 gp41 in lipid detergent and aqueous environments using 13c-3 enhanced fourier transform infrared spectroscopy
19 c5xtdo_
7.5
16
PDB header: oxidoreductase/electron transportChain: O: PDB Molecule: nadh dehydrogenase [ubiquinone] flavoprotein 2,PDBTitle: cryo-em structure of human respiratory complex i
20 c1s0bA_
7.4
33
PDB header: toxin, hydrolaseChain: A: PDB Molecule: botulinum neurotoxin type b;PDBTitle: crystal structure of botulinum neurotoxin type b at ph 4.0
21 c4pv1E_
not modelled
7.2
12
PDB header: electron transport/inhibitorChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: cytochrome b6f structure from m. laminosus with the quinone analog2 inhibitor stigmatellin
22 c4i7zE_
not modelled
7.2
12
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of cytochrome b6f in dopg, with disordered rieske2 iron-sulfur protein soluble domain
23 c5vf3Z_
not modelled
6.4
31
PDB header: virusChain: Z: PDB Molecule: highly immunogenic outer capsid protein;PDBTitle: bacteriophage t4 isometric capsid
24 c5ldwg_
not modelled
6.4
18
PDB header: oxidoreductaseChain: G: PDB Molecule: nadh-ubiquinone oxidoreductase 75 kda subunit,PDBTitle: structure of mammalian respiratory complex i, class1
25 c6b4iD_
not modelled
6.3
20
PDB header: transport proteinChain: D: PDB Molecule: nucleoporin like 2;PDBTitle: crystal structure of human gle1 ctd-nup42 gbm-ddx19b(adp) complex
26 d3btaa2
not modelled
6.3
17
Fold: beta-TrefoilSuperfamily: STI-likeFamily: Clostridium neurotoxins, C-terminal domain
27 c6hu9t_
not modelled
6.2
23
PDB header: oxidoreductase/electron transportChain: T: PDB Molecule: cytochrome b-c1 complex subunit 9;PDBTitle: iii2-iv2 mitochondrial respiratory supercomplex from s. cerevisiae
28 c3jcuw_
not modelled
6.1
20
PDB header: membrane proteinChain: W: PDB Molecule: photosystem ii reaction center w protein, chloroplastic;PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
29 d1kf6c_
not modelled
6.0
9
Fold: Heme-binding four-helical bundleSuperfamily: Fumarate reductase respiratory complex transmembrane subunitsFamily: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)
30 c6b4jD_
not modelled
5.9
20
PDB header: transport proteinChain: D: PDB Molecule: nucleoporin like 2;PDBTitle: crystal structure of human gle1 ctd-nup42 gbm-ddx19b(amppnp) complex
31 d3pcca_
not modelled
5.9
35
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase
32 c2e74E_
not modelled
5.7
12
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex from m.laminosus
33 c4h13E_
not modelled
5.7
12
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex from mastigocladus2 laminosus with tds
34 c2e76E_
not modelled
5.7
12
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
35 d2e74e1
not modelled
5.7
12
Fold: Single transmembrane helixSuperfamily: PetL subunit of the cytochrome b6f complexFamily: PetL subunit of the cytochrome b6f complex
36 c2e75E_
not modelled
5.7
12
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex with 2-nonyl-4-2 hydroxyquinoline n-oxide (nqno) from m.laminosus
37 c4h0lE_
not modelled
5.7
12
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: cytochrome b6f complex crystal structure from mastigocladus laminosus2 with n-side inhibitor nqno
38 c1vf5R_
not modelled
5.7
12
PDB header: photosynthesisChain: R: PDB Molecule: protein pet l;PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
39 d2burb1
not modelled
5.6
35
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase
40 d3pccm_
not modelled
5.5
29
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase
41 c4l6uB_
not modelled
5.5
33
PDB header: unknown functionChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of af1868: cmr1 subunit of the cmr rna silencing2 complex
42 d2j8cl1
not modelled
5.4
30
Fold: Bacterial photosystem II reaction centre, L and M subunitsSuperfamily: Bacterial photosystem II reaction centre, L and M subunitsFamily: Bacterial photosystem II reaction centre, L and M subunits
43 c2boyC_
not modelled
5.4
47
PDB header: oxidoreductaseChain: C: PDB Molecule: 3-chlorocatechol 1,2-dioxygenase;PDBTitle: crystal structure of 3-chlorocatechol 1,2-dioxygenase from rhodococcus2 opacus 1cp