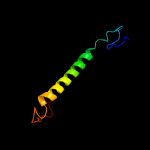
1 c5ir6B_
100.0
17
PDB header: oxidoreductaseChain: B: PDB Molecule: bd-type quinol oxidase subunit ii;PDBTitle: the structure of bd oxidase from geobacillus thermodenitrificans
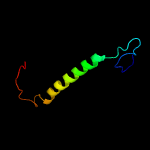
2 c6cfwG_
87.9
10
PDB header: membrane proteinChain: G: PDB Molecule: monovalent cation/h+ antiporter subunit c;PDBTitle: cryoem structure of a respiratory membrane-bound hydrogenase
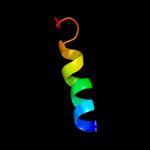
3 c6f0kC_
71.6
10
PDB header: membrane proteinChain: C: PDB Molecule: polysulphide reductase nrfd;PDBTitle: alternative complex iii
4 c3m7eA_
57.5
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: tellurite resistance protein teha homolog;PDBTitle: crystal structure of plant slac1 homolog teha
5 c3m7bA_
57.5
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: tellurite resistance protein teha homolog;PDBTitle: crystal structure of plant slac1 homolog teha
6 c2yevB_
45.1
14
PDB header: electron transportChain: B: PDB Molecule: cytochrome c oxidase subunit 2;PDBTitle: structure of caa3-type cytochrome oxidase
7 c4djiA_
35.3
15
PDB header: transport proteinChain: A: PDB Molecule: probable glutamate/gamma-aminobutyrate antiporter;PDBTitle: structure of glutamate-gaba antiporter gadc
8 d1fftb2
34.1
19
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region
9 c5ir6A_
29.3
14
PDB header: oxidoreductaseChain: A: PDB Molecule: bd-type quinol oxidase subunit i;PDBTitle: the structure of bd oxidase from geobacillus thermodenitrificans
10 c6f0kF_
26.6
12
PDB header: membrane proteinChain: F: PDB Molecule: actf;PDBTitle: alternative complex iii
11 d1ejxc2
24.2
25
Fold: TIM beta/alpha-barrelSuperfamily: Metallo-dependent hydrolasesFamily: alpha-subunit of urease, catalytic domain
12 c5wudA_
24.1
28
PDB header: membrane proteinChain: A: PDB Molecule: uncharacterized protein;PDBTitle: structural basis for conductance through tric cation channels
13 d1e9yb2
22.3
22
Fold: TIM beta/alpha-barrelSuperfamily: Metallo-dependent hydrolasesFamily: alpha-subunit of urease, catalytic domain
14 c4lhdB_
18.9
38
PDB header: oxidoreductaseChain: B: PDB Molecule: glycine dehydrogenase [decarboxylating];PDBTitle: crystal structure of synechocystis sp. pcc 6803 glycine decarboxylase2 (p-protein), holo form with pyridoxal-5'-phosphate and glycine,3 closed flexible loop
15 c4rymA_
18.1
11
PDB header: membrane proteinChain: A: PDB Molecule: integral membrane protein;PDBTitle: crystal structure of bctspo iodo type1 monomer
16 d2r6gf1
17.7
13
Fold: MalF N-terminal region-likeSuperfamily: MalF N-terminal region-likeFamily: MalF N-terminal region-like
17 c1ar1B_
16.6
9
PDB header: complex (oxidoreductase/antibody)Chain: B: PDB Molecule: cytochrome c oxidase;PDBTitle: structure at 2.7 angstrom resolution of the paracoccus2 denitrificans two-subunit cytochrome c oxidase complexed3 with an antibody fv fragment
18 c1qleB_
16.6
9
PDB header: oxidoreductase/immune systemChain: B: PDB Molecule: cytochrome c oxidase polypeptide ii;PDBTitle: cryo-structure of the paracoccus denitrificans four-subunit cytochrome2 c oxidase in the completely oxidized state complexed with an antibody3 fv fragment
19 c5fayA_
14.9
19
PDB header: lyaseChain: A: PDB Molecule: choline trimethylamine-lyase;PDBTitle: y208f mutant of choline tma-lyase
20 c2bo9B_
11.4
19
PDB header: hydrolaseChain: B: PDB Molecule: human latexin;PDBTitle: human carboxypeptidase a4 in complex with human latexin.
21 c2f3oB_
not modelled
10.7
19
PDB header: unknown functionChain: B: PDB Molecule: pyruvate formate-lyase 2;PDBTitle: crystal structure of a glycyl radical enzyme from archaeoglobus2 fulgidus
22 c5a0uA_
not modelled
10.6
29
PDB header: lyaseChain: A: PDB Molecule: choline trimethylamine lyase;PDBTitle: structure of cutc choline lyase choline bound form from klebsiella2 pneumoniae.
23 c5ymrB_
not modelled
10.3
19
PDB header: lyaseChain: B: PDB Molecule: formate acetyltransferase;PDBTitle: the crystal structure of iseg
24 c6hwhL_
not modelled
10.3
12
PDB header: electron transportChain: L: PDB Molecule: cytochrome c oxidase subunit 2;PDBTitle: structure of a functional obligate respiratory supercomplex from2 mycobacterium smegmatis
25 c5lnkn_
not modelled
8.9
24
PDB header: oxidoreductaseChain: N: PDB Molecule: mitochondrial complex i, nd2 subunit;PDBTitle: entire ovine respiratory complex i
26 c5wufA_
not modelled
8.1
23
PDB header: membrane proteinChain: A: PDB Molecule: putative membrane protein;PDBTitle: structural basis for conductance through tric cation channels
27 c2ktaA_
not modelled
7.9
42
PDB header: hydrolaseChain: A: PDB Molecule: putative helicase;PDBTitle: solution nmr structure of a domain of protein a6ky75 from bacteroides2 vulgatus, northeast structural genomics target bvr106a
28 c5xmjG_
not modelled
7.8
14
PDB header: electron transportChain: G: PDB Molecule: fumarate reductase respiratory complex;PDBTitle: crystal structure of quinol:fumarate reductase from desulfovibrio2 gigas
29 c5h36E_
not modelled
7.7
17
PDB header: membrane proteinChain: E: PDB Molecule: uncharacterized protein tric;PDBTitle: crystal structures of the tric trimeric intracellular cation channel2 orthologue from rhodobacter sphaeroides
30 c1fftG_
not modelled
7.6
8
PDB header: oxidoreductaseChain: G: PDB Molecule: ubiquinol oxidase;PDBTitle: the structure of ubiquinol oxidase from escherichia coli
31 c5h35C_
not modelled
7.6
16
PDB header: immune system/membrane proteinChain: C: PDB Molecule: membrane protein tric;PDBTitle: crystal structures of the tric trimeric intracellular cation channel2 orthologue from sulfolobus solfataricus
32 c5i1mV_
not modelled
7.2
13
PDB header: membrane proteinChain: V: PDB Molecule: v-type proton atpase subunit a, vacuolar isoform;PDBTitle: yeast v-atpase average of densities, a subunit segment
33 c5jffC_
not modelled
7.2
27
PDB header: transferaseChain: C: PDB Molecule: probable adenosine monophosphate-protein transferase fic;PDBTitle: e. coli ecfict mutant g55r in complex with ecfica
34 c4x2eA_
not modelled
7.2
36
PDB header: transferaseChain: A: PDB Molecule: fic family protein putative filamentation induced by campPDBTitle: clostridium difficile wild type fic protein
35 c4ylfA_
not modelled
7.2
60
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydroorotate dehydrogenase b (nad(+)), electron transferPDBTitle: insights into flavin-based electron bifurcation via the nadh-dependent2 reduced ferredoxin-nadp oxidoreductase structure
36 c5ly0B_
not modelled
6.9
18
PDB header: transcriptionChain: B: PDB Molecule: lob family transfactor ramosa2.1;PDBTitle: crystal structure of lob domain of ramosa2 from wheat
37 c4heaJ_
not modelled
6.7
9
PDB header: oxidoreductaseChain: J: PDB Molecule: nadh-quinone oxidoreductase subunit 10;PDBTitle: crystal structure of the entire respiratory complex i from thermus2 thermophilus
38 c4hkrB_
not modelled
6.4
14
PDB header: transport proteinChain: B: PDB Molecule: calcium release-activated calcium channel protein 1;PDBTitle: calcium release-activated calcium (crac) channel orai
39 c2mgyA_
not modelled
5.9
20
PDB header: membrane proteinChain: A: PDB Molecule: translocator protein;PDBTitle: solution structure of the mitochondrial translocator protein (tspo) in2 complex with its high-affinity ligand pk11195
40 c2hfzA_
not modelled
5.5
23
PDB header: transferaseChain: A: PDB Molecule: rna-directed rna polymerase(ns5);PDBTitle: crystal structure of rna dependent rna polymerase domain2 from west nile virus
41 c5voxb_
not modelled
5.5
14
PDB header: hydrolaseChain: B: PDB Molecule: v-type proton atpase subunit b;PDBTitle: yeast v-atpase in complex with legionella pneumophila effector sidk2 (rotational state 1)
42 c2v5iA_
not modelled
5.3
63
PDB header: viral proteinChain: A: PDB Molecule: salmonella typhimurium db7155 bacteriophage det7 tailspike;PDBTitle: structure of the receptor-binding protein of bacteriophage det7: a2 podoviral tailspike in a myovirus
43 c4hkrA_
not modelled
5.2
14
PDB header: transport proteinChain: A: PDB Molecule: calcium release-activated calcium channel protein 1;PDBTitle: calcium release-activated calcium (crac) channel orai
44 c6nbxG_
not modelled
5.2
14
PDB header: oxidoreductaseChain: G: PDB Molecule: nadh-quinone oxidoreductase subunit j;PDBTitle: t.elongatus ndh (data-set 2)