| 1 |
|
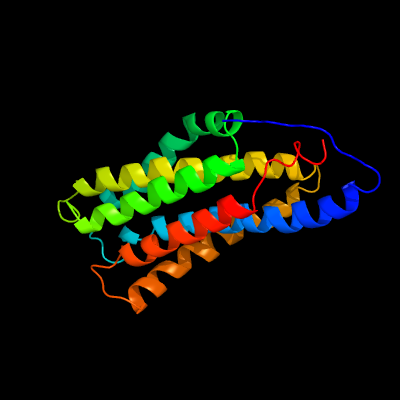
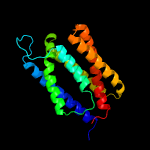
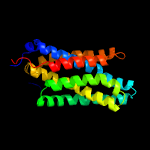
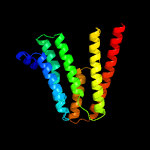
PDB 1pw4 chain A
Region: 4 - 195
Aligned: 192
Modelled: 192
Confidence: 73.2%
Identity: 10%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 2 |
|
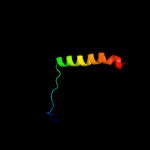
PDB 6bas chain A
Region: 21 - 177
Aligned: 146
Modelled: 157
Confidence: 31.9%
Identity: 11%
PDB header:transferase
Chain: A: PDB Molecule:peptidoglycan glycosyltransferase roda;
PDBTitle: crystal structure of thermus thermophilus rod shape determining2 protein roda d255a mutant (q5six3_thet8)
Phyre2
| 3 |
|
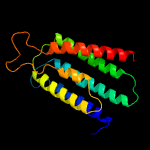
PDB 4pgu chain A
Region: 13 - 193
Aligned: 175
Modelled: 181
Confidence: 25.7%
Identity: 12%
PDB header:membrance protein
Chain: A: PDB Molecule:uncharacterized protein yetj;
PDBTitle: crystal structure of yetj from bacillus subtilis at ph 7 by soaking
Phyre2
| 4 |
|
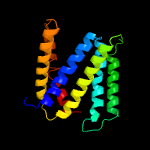
PDB 3arc chain L
Region: 7 - 42
Aligned: 36
Modelled: 36
Confidence: 24.7%
Identity: 25%
PDB header:electron transport, photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
Phyre2
| 5 |
|
PDB 3j1z chain P
Region: 18 - 187
Aligned: 170
Modelled: 170
Confidence: 20.7%
Identity: 10%
PDB header:metal transport
Chain: P: PDB Molecule:cation efflux family protein;
PDBTitle: inward-facing conformation of the zinc transporter yiip revealed by2 cryo-electron microscopy
Phyre2
| 6 |
|
PDB 5i20 chain C
Region: 21 - 191
Aligned: 171
Modelled: 171
Confidence: 15.4%
Identity: 11%
PDB header:membrane protein
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of protein
Phyre2
| 7 |
|
PDB 1h59 chain B
Region: 2 - 16
Aligned: 15
Modelled: 15
Confidence: 14.9%
Identity: 27%
Fold: Knottins (small inhibitors, toxins, lectins)
Superfamily: Growth factor receptor domain
Family: Growth factor receptor domain
Phyre2
| 8 |
|
PDB 6bm8 chain A
Region: 142 - 176
Aligned: 35
Modelled: 35
Confidence: 14.7%
Identity: 23%
PDB header:viral protein
Chain: A: PDB Molecule:envelope glycoprotein b;
PDBTitle: crystal structure of glycoprotein b from herpes simplex virus type i
Phyre2
| 9 |
|
PDB 6btm chain C
Region: 9 - 193
Aligned: 185
Modelled: 185
Confidence: 13.6%
Identity: 15%
PDB header:membrane protein
Chain: C: PDB Molecule:alternative complex iii subunit c;
PDBTitle: structure of alternative complex iii from flavobacterium johnsoniae2 (wild type)
Phyre2
| 10 |
|
PDB 5i20 chain E
Region: 21 - 195
Aligned: 172
Modelled: 175
Confidence: 13.4%
Identity: 14%
PDB header:membrane protein
Chain: E: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of protein
Phyre2
| 11 |
|
PDB 6g9x chain B
Region: 1 - 193
Aligned: 193
Modelled: 193
Confidence: 11.9%
Identity: 14%
PDB header:membrane protein
Chain: B: PDB Molecule:major facilitator superfamily mfs_1;
PDBTitle: crystal structure of a mfs transporter at 2.54 angstroem resolution
Phyre2
| 12 |
|
PDB 2ide chain E
Region: 1 - 13
Aligned: 13
Modelled: 13
Confidence: 9.2%
Identity: 31%
PDB header:biosynthetic protein
Chain: E: PDB Molecule:molybdenum cofactor biosynthesis protein c;
PDBTitle: crystal structure of the molybdenum cofactor biosynthesis protein c2 (ttha1789) from thermus theromophilus hb8
Phyre2
| 13 |
|
PDB 1ekr chain A
Region: 1 - 13
Aligned: 13
Modelled: 13
Confidence: 8.8%
Identity: 23%
Fold: Ferredoxin-like
Superfamily: Molybdenum cofactor biosynthesis protein C, MoaC
Family: Molybdenum cofactor biosynthesis protein C, MoaC
Phyre2
| 14 |
|
PDB 5wlb chain C
Region: 6 - 24
Aligned: 19
Modelled: 19
Confidence: 8.2%
Identity: 26%
PDB header:protein binding
Chain: C: PDB Molecule:225-15 b;
PDBTitle: kras g12v, bound to gppnhp and miniprotein 225-15a/b
Phyre2
| 15 |
|
PDB 2ohd chain B
Region: 1 - 13
Aligned: 13
Modelled: 13
Confidence: 7.7%
Identity: 46%
PDB header:biosynthetic protein
Chain: B: PDB Molecule:probable molybdenum cofactor biosynthesis protein c;
PDBTitle: crystal structure of hypothetical molybdenum cofactor biosynthesis2 protein c from sulfolobus tokodaii
Phyre2
| 16 |
|
PDB 2eey chain A
Region: 1 - 13
Aligned: 13
Modelled: 13
Confidence: 7.7%
Identity: 23%
PDB header:biosynthetic protein
Chain: A: PDB Molecule:molybdopterin biosynthesis;
PDBTitle: structure of gk0241 protein from geobacillus kaustophilus
Phyre2
| 17 |
|
PDB 3ue5 chain B
Region: 1 - 6
Aligned: 6
Modelled: 6
Confidence: 7.3%
Identity: 50%
PDB header:contractile protein/transport protein
Chain: B: PDB Molecule:protein spire;
PDBTitle: ecp-cleaved actin in complex with spir domain d
Phyre2
| 18 |
|
PDB 4fdf chain B
Region: 1 - 13
Aligned: 13
Modelled: 13
Confidence: 6.2%
Identity: 23%
PDB header:biosynthetic protein
Chain: B: PDB Molecule:molybdenum cofactor biosynthesis protein c 2;
PDBTitle: structural insights into putative molybdenum cofactor biosynthesis2 protein c (moac2) from mycobacterium tuberculosis h37rv
Phyre2
| 19 |
|
PDB 1r89 chain A domain 3
Region: 3 - 18
Aligned: 16
Modelled: 16
Confidence: 5.9%
Identity: 19%
Fold: Ferredoxin-like
Superfamily: PAP/Archaeal CCA-adding enzyme, C-terminal domain
Family: Archaeal tRNA CCA-adding enzyme
Phyre2
| 20 |
|
PDB 5d92 chain B
Region: 10 - 190
Aligned: 181
Modelled: 181
Confidence: 5.4%
Identity: 14%
PDB header:membrane protein
Chain: B: PDB Molecule:af2299 protein,phosphatidylinositol synthase;
PDBTitle: structure of a phosphatidylinositolphosphate (pip) synthase from2 renibacterium salmoninarum
Phyre2
| 21 |
|