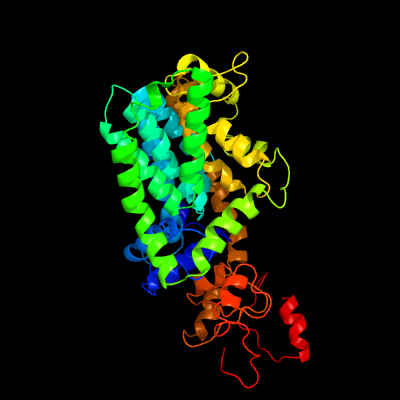
| 1 | c5f15A_
|
|
 |
100.0 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:4-amino-4-deoxy-l-arabinose (l-ara4n) transferase;
PDBTitle: crystal structure of arnt from cupriavidus metallidurans bound to2 undecaprenyl phosphate
|
|
|
|
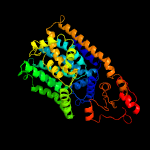
| 2 | c6p2rB_
|
|
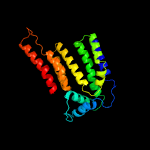 |
99.8 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:dolichyl-phosphate-mannose--protein mannosyltransferase 2;
PDBTitle: structure of s. cerevisiae protein o-mannosyltransferase pmt1-pmt22 complex bound to the sugar donor
|
|
|
|
| 3 | c6p25A_
|
|
 |
99.8 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:dolichyl-phosphate-mannose--protein mannosyltransferase 1;
PDBTitle: structure of s. cerevisiae protein o-mannosyltransferase pmt1-pmt22 complex bound to the sugar donor and a peptide acceptor
|
|
|
|
| 4 | c3wajA_
|
|
 |
99.7 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:transmembrane oligosaccharyl transferase;
PDBTitle: crystal structure of the archaeoglobus fulgidus2 oligosaccharyltransferase (o29867_arcfu) complex with zn and sulfate
|
|
|
|
| 5 | c3rceA_
|
|
 |
99.7 |
12 |
PDB header:transferase/peptide
Chain: A: PDB Molecule:oligosaccharide transferase to n-glycosylate proteins;
PDBTitle: bacterial oligosaccharyltransferase pglb
|
|
|
|
| 6 | c6eznF_
|
|
 |
99.5 |
16 |
PDB header:membrane protein
Chain: F: PDB Molecule:dolichyl-diphosphooligosaccharide--protein
PDBTitle: cryo-em structure of the yeast oligosaccharyltransferase (ost) complex
|
|
|
|
| 7 | d2jfga2
|
|
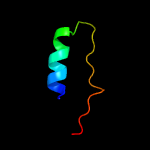 |
79.5 |
21 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
|
|
|
| 8 | c3uagA_
|
|
 |
42.3 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:protein (udp-n-acetylmuramoyl-l-alanine:d-
PDBTitle: udp-n-acetylmuramoyl-l-alanine:d-glutamate ligase
|
|
|
|
| 9 | c3lk7A_
|
|
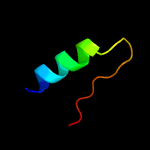 |
37.3 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramoylalanine--d-glutamate ligase;
PDBTitle: the crystal structure of udp-n-acetylmuramoylalanine-d-glutamate2 (murd) ligase from streptococcus agalactiae to 1.5a
|
|
|
|
| 10 | c3mvnA_
|
|
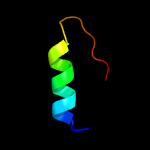 |
29.3 |
32 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate:l-alanyl-gamma-d-glutamyl-medo-
PDBTitle: crystal structure of a domain from a putative udp-n-acetylmuramate:l-2 alanyl-gamma-d-glutamyl-medo-diaminopimelate ligase from haemophilus3 ducreyi 35000hp
|
|
|
|
| 11 | d1xq4a_
|
|
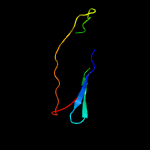 |
27.7 |
21 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:ApaG-like
Family:ApaG-like |
|
|
|
| 12 | c2f1eA_
|
|
 |
27.5 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein apag;
PDBTitle: solution structure of apag protein
|
|
|
|
| 13 | c5hdwA_
|
|
 |
26.0 |
24 |
PDB header:protein binding
Chain: A: PDB Molecule:f-box only protein 3;
PDBTitle: apag domain of fbxo3
|
|
|
|
| 14 | d1tzaa_
|
|
 |
25.7 |
16 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:ApaG-like
Family:ApaG-like |
|
|
|
| 15 | d1xvsa_
|
|
 |
25.0 |
9 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:ApaG-like
Family:ApaG-like |
|
|
|
| 16 | c5n6mA_
|
|
 |
19.9 |
13 |
PDB header:membrane protein
Chain: A: PDB Molecule:apolipoprotein n-acyltransferase;
PDBTitle: structure of the membrane integral lipoprotein n-acyltransferase lnt2 from p. aeruginosa
|
|
|
|
| 17 | d1p3da2
|
|
 |
18.7 |
36 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
|
|
|
| 18 | c3hn7A_
|
|
 |
18.6 |
27 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate-l-alanine ligase;
PDBTitle: crystal structure of a murein peptide ligase mpl (psyc_0032) from2 psychrobacter arcticus 273-4 at 1.65 a resolution
|
|
|
|
| 19 | c4bucA_
|
|
 |
18.1 |
13 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramoylalanine--d-glutamate ligase;
PDBTitle: crystal structure of murd ligase from thermotoga maritima in apo form
|
|
|
|
| 20 | d1e8ca2
|
|
 |
14.5 |
23 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
|
|
|
| 21 | c3tr7A_ |
|
not modelled |
14.3 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:uracil-dna glycosylase;
PDBTitle: structure of a uracil-dna glycosylase (ung) from coxiella burnetii
|
|
|
| 22 | c6nbxG_ |
|
not modelled |
12.9 |
14 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:nadh-quinone oxidoreductase subunit j;
PDBTitle: t.elongatus ndh (data-set 2)
|
|
|
| 23 | d1vk3a3 |
|
not modelled |
11.8 |
18 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 24 | c3gt2A_ |
|
not modelled |
11.3 |
18 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the p60 domain from m. avium paratuberculosis2 antigen map1272c
|
|
|
| 25 | c4c13A_ |
|
not modelled |
11.2 |
23 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramoyl-l-alanyl-d-glutamate--l-lysine ligase;
PDBTitle: x-ray crystal structure of staphylococcus aureus mure with udp-murnac-2 ala-glu-lys
|
|
|
| 26 | d1h9aa1 |
|
not modelled |
11.1 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 27 | c2fynH_ |
|
not modelled |
10.9 |
14 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:cytochrome c1;
PDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
|
|
|
| 28 | c1e8cB_ |
|
not modelled |
10.7 |
23 |
PDB header:ligase
Chain: B: PDB Molecule:udp-n-acetylmuramoylalanyl-d-glutamate--2,6-diaminopimelate
PDBTitle: structure of mure the udp-n-acetylmuramyl tripeptide synthetase from2 e. coli
|
|
|
| 29 | d1s04a_ |
|
not modelled |
10.7 |
27 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:ProFAR isomerase associated domain |
|
|
| 30 | c1zrtD_ |
|
not modelled |
10.6 |
11 |
PDB header:oxidoreductase/metal transport
Chain: D: PDB Molecule:cytochrome c1;
PDBTitle: rhodobacter capsulatus cytochrome bc1 complex with2 stigmatellin bound
|
|
|
| 31 | c2f00A_ |
|
not modelled |
10.6 |
27 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate--l-alanine ligase;
PDBTitle: escherichia coli murc
|
|
|
| 32 | d2evra2 |
|
not modelled |
10.5 |
21 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:NlpC/P60 |
|
|
| 33 | c4afpA_ |
|
not modelled |
10.1 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:metacaspase mca2;
PDBTitle: the structure of metacaspase 2 from t. brucei determined in the2 presence of samarium
|
|
|
| 34 | c5ifiA_ |
|
not modelled |
10.0 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:acetyl-coenzyme a synthetase;
PDBTitle: crystal structure of acetyl-coa synthetase in complex with adenosine-2 5'-propylphosphate from cryptococcus neoformans h99
|
|
|
| 35 | c6b8cA_ |
|
not modelled |
10.0 |
36 |
PDB header:hydrolase
Chain: A: PDB Molecule:nlp/p60;
PDBTitle: crystal structure of nlpc/p60 domain of peptidoglycan hydrolase saga
|
|
|
| 36 | c4bubA_ |
|
not modelled |
9.9 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramoyl-l-alanyl-d-glutamate--ld-lysine
PDBTitle: crystal structure of mure ligase from thermotoga maritima2 in complex with adp
|
|
|
| 37 | c1gqqA_ |
|
not modelled |
9.8 |
36 |
PDB header:cell wall biosynthesis
Chain: A: PDB Molecule:udp-n-acetylmuramate-l-alanine ligase;
PDBTitle: murc - crystal structure of the apo-enzyme from haemophilus influenzae
|
|
|
| 38 | d1vkya_ |
|
not modelled |
9.8 |
31 |
Fold:QueA-like
Superfamily:QueA-like
Family:QueA-like |
|
|
| 39 | c4uy5A_ |
|
not modelled |
9.7 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:histidine-specific methyltransferase egtd;
PDBTitle: crystal structure of histidine-specific methyltransferase egtd from2 mycobacterium smegmatis
|
|
|
| 40 | c2yiuE_ |
|
not modelled |
9.7 |
11 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:cytochrome c1, heme protein;
PDBTitle: x-ray structure of the dimeric cytochrome bc1 complex from2 the soil bacterium paracoccus denitrificans at 2.73 angstrom resolution
|
|
|
| 41 | d1ofla_ |
|
not modelled |
9.6 |
17 |
Fold:Single-stranded right-handed beta-helix
Superfamily:Pectin lyase-like
Family:Chondroitinase B |
|
|
| 42 | c2lkjA_ |
|
not modelled |
9.5 |
50 |
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-m;
PDBTitle: structures and interaction analyses of the integrin alpha-m beta-22 cytoplasmic tails
|
|
|
| 43 | c5eqnA_ |
|
not modelled |
9.3 |
50 |
PDB header:hydrolase
Chain: A: PDB Molecule:frbj;
PDBTitle: structure of phosphonate hydroxylase
|
|
|
| 44 | d3c9ua2 |
|
not modelled |
9.3 |
33 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 45 | d1jr7a_ |
|
not modelled |
9.2 |
33 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Gab protein (hypothetical protein YgaT) |
|
|
| 46 | c3n2aA_ |
|
not modelled |
9.2 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:bifunctional folylpolyglutamate synthase/dihydrofolate
PDBTitle: crystal structure of bifunctional folylpolyglutamate2 synthase/dihydrofolate synthase from yersinia pestis co92
|
|
|
| 47 | c5ey8D_ |
|
not modelled |
9.2 |
18 |
PDB header:ligase
Chain: D: PDB Molecule:acyl-coa synthase;
PDBTitle: structure of fadd32 from mycobacterium smegmatis complexed to ampc20
|
|
|
| 48 | d1wdia_ |
|
not modelled |
9.2 |
25 |
Fold:QueA-like
Superfamily:QueA-like
Family:QueA-like |
|
|
| 49 | c3cwbQ_ |
|
not modelled |
9.1 |
8 |
PDB header:oxidoreductase
Chain: Q: PDB Molecule:mitochondrial cytochrome c1, heme protein;
PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
|
|
|
| 50 | c1yy3A_ |
|
not modelled |
9.1 |
44 |
PDB header:isomerase
Chain: A: PDB Molecule:s-adenosylmethionine:trna ribosyltransferase-
PDBTitle: structure of s-adenosylmethionine:trna ribosyltransferase-2 isomerase (quea)
|
|
|
| 51 | c4e9iB_ |
|
not modelled |
9.1 |
33 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glucose-6-phosphate 1-dehydrogenase;
PDBTitle: glucose-6-p dehydrogenase (apo form) from trypanosoma cruzi
|
|
|
| 52 | c3on0D_ |
|
not modelled |
9.0 |
18 |
PDB header:dna binding protein/dna
Chain: D: PDB Molecule:protein tram;
PDBTitle: crystal structure of the ped208 tram-sbma complex
|
|
|
| 53 | c3etcB_ |
|
not modelled |
8.9 |
10 |
PDB header:ligase
Chain: B: PDB Molecule:amp-binding protein;
PDBTitle: 2.1 a structure of acyl-adenylate synthetase from methanosarcina2 acetivorans containing a link between lys256 and cys298
|
|
|
| 54 | c1qkiE_ |
|
not modelled |
8.9 |
33 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:glucose-6-phosphate 1-dehydrogenase;
PDBTitle: x-ray structure of human glucose 6-phosphate dehydrogenase (variant2 canton r459l) complexed with structural nadp+
|
|
|
| 55 | c3rg2H_ |
|
not modelled |
8.8 |
15 |
PDB header:ligase
Chain: H: PDB Molecule:enterobactin synthase component e (ente), 2,3-dihydro-2,3-
PDBTitle: structure of a two-domain nrps fusion protein containing the ente2 adenylation domain and entb aryl-carrier protein from enterobactin3 biosynthesis
|
|
|
| 56 | c1qcrD_ |
|
not modelled |
8.7 |
5 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:ubiquinol cytochrome c oxidoreductase;
PDBTitle: crystal structure of bovine mitochondrial cytochrome bc12 complex, alpha carbon atoms only
|
|
|
| 57 | d2zoda2 |
|
not modelled |
8.7 |
25 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 58 | c1w78A_ |
|
not modelled |
8.7 |
36 |
PDB header:synthase
Chain: A: PDB Molecule:folc bifunctional protein;
PDBTitle: e.coli folc in complex with dhpp and adp
|
|
|
| 59 | d1xnea_ |
|
not modelled |
8.6 |
20 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:ProFAR isomerase associated domain |
|
|
| 60 | c1dbgA_ |
|
not modelled |
8.5 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:chondroitinase b;
PDBTitle: crystal structure of chondroitinase b
|
|
|
| 61 | c2bhlB_ |
|
not modelled |
8.5 |
33 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glucose-6-phosphate 1-dehydrogenase;
PDBTitle: x-ray structure of human glucose-6-phosphate dehydrogenase (deletion2 variant) complexed with glucose-6-phosphate
|
|
|
| 62 | c2xivA_ |
|
not modelled |
8.5 |
12 |
PDB header:structural protein
Chain: A: PDB Molecule:hypothetical invasion protein;
PDBTitle: structure of rv1477, hypothetical invasion protein of mycobacterium2 tuberculosis
|
|
|
| 63 | d2pyta1 |
|
not modelled |
8.3 |
24 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:EutQ-like |
|
|
| 64 | d1t33a2 |
|
not modelled |
8.1 |
12 |
Fold:Tetracyclin repressor-like, C-terminal domain
Superfamily:Tetracyclin repressor-like, C-terminal domain
Family:Tetracyclin repressor-like, C-terminal domain |
|
|
| 65 | c2xjaD_ |
|
not modelled |
8.1 |
32 |
PDB header:ligase
Chain: D: PDB Molecule:udp-n-acetylmuramoyl-l-alanyl-d-glutamate--2,6-
PDBTitle: structure of mure from m.tuberculosis with dipeptide and adp
|
|
|
| 66 | c1h9aA_ |
|
not modelled |
8.1 |
29 |
PDB header:oxidoreductase (choh(d) - nad(p))
Chain: A: PDB Molecule:glucose 6-phosphate 1-dehydrogenase;
PDBTitle: complex of active mutant (q365->c) of glucose 6-phosphate2 dehydrogenase from l. mesenteroides with coenzyme nadp
|
|
|
| 67 | c4r0mA_ |
|
not modelled |
8.0 |
10 |
PDB header:ligase
Chain: A: PDB Molecule:mcyg protein;
PDBTitle: structure of mcyg a-pcp complexed with phenylalanyl-adenylate
|
|
|
| 68 | d1pg4a_ |
|
not modelled |
7.9 |
16 |
Fold:Acetyl-CoA synthetase-like
Superfamily:Acetyl-CoA synthetase-like
Family:Acetyl-CoA synthetase-like |
|
|
| 69 | c6ijbA_ |
|
not modelled |
7.9 |
9 |
PDB header:ligase
Chain: A: PDB Molecule:amp-binding domain protein;
PDBTitle: structure of 3-methylmercaptopropionate coa ligase mutant k523a in2 complex with amp and mmpa
|
|
|
| 70 | c1p84D_ |
|
not modelled |
7.7 |
8 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:cytochrome c1, heme protein;
PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
|
|
|
| 71 | c3rkoF_ |
|
not modelled |
7.7 |
11 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:nadh-quinone oxidoreductase subunit j;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
|
|
|
| 72 | c4cmrB_ |
|
not modelled |
7.7 |
41 |
PDB header:hydrolase
Chain: B: PDB Molecule:glycosyl hydrolase/deacetylase family protein;
PDBTitle: the crystal structure of novel exo-type maltose-forming2 amylase(py04_0872) from pyrococcus sp. st04
|
|
|
| 73 | d3euga_ |
|
not modelled |
7.5 |
12 |
Fold:Uracil-DNA glycosylase-like
Superfamily:Uracil-DNA glycosylase-like
Family:Uracil-DNA glycosylase |
|
|
| 74 | d1iz0a1 |
|
not modelled |
7.5 |
5 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
|
|
| 75 | c5mvrA_ |
|
not modelled |
7.4 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:trna threonylcarbamoyladenosine biosynthesis protein tsae;
PDBTitle: crystal structure of bacillus subtilus ydib
|
|
|
| 76 | c4gr5B_ |
|
not modelled |
7.4 |
11 |
PDB header:ligase
Chain: B: PDB Molecule:non-ribosomal peptide synthetase;
PDBTitle: crystal structure of slgn1deltaasub in complex with ampcpp
|
|
|
| 77 | c6biqA_ |
|
not modelled |
7.4 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:clan ca, family c40, nlpc/p60 superfamily cysteine
PDBTitle: structure of nlpc2 from trichomonas vaginalis
|
|
|
| 78 | d1clia2 |
|
not modelled |
7.4 |
27 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 79 | c3ibmB_ |
|
not modelled |
7.3 |
15 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:cupin 2, conserved barrel domain protein;
PDBTitle: crystal structure of cupin 2 domain-containing protein hhal_0468 from2 halorhodospira halophila
|
|
|
| 80 | d2z1ea2 |
|
not modelled |
7.3 |
53 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 81 | c2wtzC_ |
|
not modelled |
7.2 |
32 |
PDB header:ligase
Chain: C: PDB Molecule:udp-n-acetylmuramoyl-l-alanyl-d-glutamate-
PDBTitle: mure ligase of mycobacterium tuberculosis
|
|
|
| 82 | c5vrhA_ |
|
not modelled |
7.1 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:apolipoprotein n-acyltransferase;
PDBTitle: apolipoprotein n-acyltransferase c387s active site mutant
|
|
|
| 83 | c2fg0B_ |
|
not modelled |
7.1 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:cog0791: cell wall-associated hydrolases (invasion-
PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (npun_r0659) from nostoc punctiforme pcc 73102 at 1.793 a resolution
|
|
|
| 84 | c5oynB_ |
|
not modelled |
7.0 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:dehydratase, ilvd/edd family;
PDBTitle: crystal structure of d-xylonate dehydratase in holo-form
|
|
|
| 85 | c4lgvA_ |
|
not modelled |
7.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glucose-6-phosphate 1-dehydrogenase;
PDBTitle: x-ray crystal structure of glucose-6-phosphate 1-dehydrogenase from2 mycobacterium avium
|
|
|
| 86 | c3fp9E_ |
|
not modelled |
7.0 |
60 |
PDB header:hydrolase
Chain: E: PDB Molecule:proteasome-associated atpase;
PDBTitle: crystal structure of intern domain of proteasome-associated2 atpase, mycobacterium tuberculosis
|
|
|
| 87 | c2k1gA_ |
|
not modelled |
7.0 |
27 |
PDB header:lipoprotein
Chain: A: PDB Molecule:lipoprotein spr;
PDBTitle: solution nmr structure of lipoprotein spr from escherichia coli k12.2 northeast structural genomics target er541-37-162
|
|
|
| 88 | c2yvaB_ |
|
not modelled |
6.8 |
29 |
PDB header:dna binding protein
Chain: B: PDB Molecule:dnaa initiator-associating protein diaa;
PDBTitle: crystal structure of escherichia coli diaa
|
|
|
| 89 | c4wd1A_ |
|
not modelled |
6.8 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:acetoacetate-coa ligase;
PDBTitle: acetoacetyl-coa synthetase from streptomyces lividans
|
|
|
| 90 | d2f4la1 |
|
not modelled |
6.8 |
50 |
Fold:CUB-like
Superfamily:Acetamidase/Formamidase-like
Family:Acetamidase/Formamidase-like |
|
|
| 91 | c5odnG_ |
|
not modelled |
6.8 |
5 |
PDB header:dna binding protein
Chain: G: PDB Molecule:single-stranded dna-binding protein;
PDBTitle: salinibacter ruber single-strand binding protein
|
|
|
| 92 | c3mjjD_ |
|
not modelled |
6.7 |
50 |
PDB header:hydrolase
Chain: D: PDB Molecule:predicted acetamidase/formamidase;
PDBTitle: crystal structure analysis of a recombinant predicted2 acetamidase/formamidase from the thermophile thermoanaerobacter3 tengcongensis
|
|
|
| 93 | c4pa5A_ |
|
not modelled |
6.7 |
32 |
PDB header:transferase
Chain: A: PDB Molecule:protein-glutamine gamma-glutamyltransferase;
PDBTitle: tgl - a bacterial spore coat transglutaminase - cystamine complex
|
|
|
| 94 | c4eatB_ |
|
not modelled |
6.7 |
14 |
PDB header:ligase
Chain: B: PDB Molecule:benzoate-coenzyme a ligase;
PDBTitle: crystal structure of a benzoate coenzyme a ligase
|
|
|
| 95 | c2ii1A_ |
|
not modelled |
6.7 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:acetamidase;
PDBTitle: crystal structure of acetamidase (10172637) from bacillus halodurans2 at 1.95 a resolution
|
|
|
| 96 | c2zw3B_ |
|
not modelled |
6.7 |
17 |
PDB header:cell adhesion
Chain: B: PDB Molecule:gap junction beta-2 protein;
PDBTitle: structure of the connexin-26 gap junction channel at 3.52 angstrom resolution
|
|
|
| 97 | c4f6oA_ |
|
not modelled |
6.7 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:metacaspase-1;
PDBTitle: crystal structure of the yeast metacaspase yca1
|
|
|
| 98 | c5ym0A_ |
|
not modelled |
6.7 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:dihydroxy-acid dehydratase, chloroplastic;
PDBTitle: the crystal structure of dhad
|
|
|
| 99 | c3kj0B_ |
|
not modelled |
6.7 |
38 |
PDB header:apoptosis
Chain: B: PDB Molecule:bcl-2-like protein 11;
PDBTitle: mcl-1 in complex with bim bh3 mutant i2dy
|
|
|