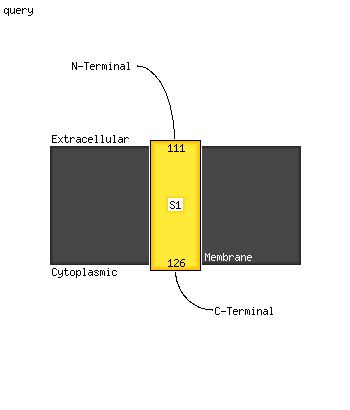
1 d1tq8a_
99.9
97
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: Universal stress protein-like
2 c5ahwC_
99.9
69
PDB header: signaling proteinChain: C: PDB Molecule: universal stress protein;PDBTitle: crystal structure of universal stress protein msmeg_3811 in2 complex with camp
3 c3s3tD_
99.9
19
PDB header: chaperoneChain: D: PDB Molecule: nucleotide-binding protein, universal stress protein uspaPDBTitle: universal stress protein uspa from lactobacillus plantarum
4 c3hgmD_
99.9
21
PDB header: signaling proteinChain: D: PDB Molecule: universal stress protein tead;PDBTitle: universal stress protein tead from the trap transporter teaabc of2 halomonas elongata
5 d2z3va1
99.9
26
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: Universal stress protein-like
6 d1mjha_
99.9
24
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: Universal stress protein-like
7 c3dloC_
99.9
27
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: universal stress protein;PDBTitle: structure of universal stress protein from archaeoglobus fulgidus
8 c3olqA_
99.9
28
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: universal stress protein e;PDBTitle: the crystal structure of a universal stress protein e from proteus2 mirabilis hi4320
9 c3fg9B_
99.9
21
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: protein of universal stress protein uspa family;PDBTitle: the crystal structure of an universal stress protein uspa2 family protein from lactobacillus plantarum wcfs1
10 d1jmva_
99.9
21
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: Universal stress protein-like
11 c4wnyA_
99.9
21
PDB header: signaling proteinChain: A: PDB Molecule: universal stress protein;PDBTitle: crystal structure of a protein from the universal stress protein2 family from burkholderia pseudomallei
12 c2dumD_
99.9
26
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: hypothetical protein ph0823;PDBTitle: crystal structure of hypothetical protein, ph0823
13 d2gm3a1
99.9
23
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: Universal stress protein-like
14 c3fh0A_
99.8
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative universal stress protein kpn_01444;PDBTitle: crystal structure of putative universal stress protein kpn_01444 -2 atpase
15 c2pfsA_
99.8
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: universal stress protein;PDBTitle: crystal structure of universal stress protein from nitrosomonas2 europaea
16 c4r2lB_
99.8
22
PDB header: unknown functionChain: B: PDB Molecule: universal stress protein f;PDBTitle: crystal structure of ynaf (universal stress protein f) from salmonella2 typhimurium
17 c4r2jA_
99.8
11
PDB header: metal binding protein, unknown functionChain: A: PDB Molecule: universal stress protein e;PDBTitle: crystal structure of ydaa (universal stress protein e) from salmonella2 typhimurium
18 c3loqA_
99.8
23
PDB header: structure genomics, unknown functionChain: A: PDB Molecule: universal stress protein;PDBTitle: the crystal structure of a universal stress protein from archaeoglobus2 fulgidus dsm 4304
19 c3idfA_
99.8
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: usp-like protein;PDBTitle: the crystal structure of a usp-like protein from wolinella2 succinogenes to 2.0a
20 d1q77a_
99.8
16
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: Universal stress protein-like
21 c3mt0A_
not modelled
99.8
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein pa1789;PDBTitle: the crystal structure of a functionally unknown protein pa1789 from2 pseudomonas aeruginosa pao1
22 c2jaxA_
not modelled
99.7
24
PDB header: protein bindingChain: A: PDB Molecule: hypothetical protein tb31.7;PDBTitle: universal stress protein rv2623 from mycobaterium2 tuberculosis
23 c3ab8B_
not modelled
99.7
26
PDB header: unknown functionChain: B: PDB Molecule: putative uncharacterized protein ttha0350;PDBTitle: crystal structure of the hypothetical tandem-type universal stress2 protein ttha0350 complexed with atps.
24 c5ol2E_
not modelled
95.2
11
PDB header: flavoproteinChain: E: PDB Molecule: electron transfer flavoprotein small subunit;PDBTitle: the electron transferring flavoprotein/butyryl-coa dehydrogenase2 complex from clostridium difficile
25 c5ow0B_
not modelled
94.0
16
PDB header: electron transportChain: B: PDB Molecule: electron transfer flavoprotein, beta subunit;PDBTitle: crystal structure of an electron transfer flavoprotein from2 geobacillus metallireducens
26 d1o94c_
not modelled
93.9
13
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits
27 d3clsc1
not modelled
93.4
12
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits
28 c6fahB_
not modelled
92.2
12
PDB header: flavoproteinChain: B: PDB Molecule: caffeyl-coa reductase-etf complex subunit card;PDBTitle: molecular basis of the flavin-based electron-bifurcating caffeyl-coa2 reductase reaction
29 c4kpuB_
not modelled
92.0
12
PDB header: electron transportChain: B: PDB Molecule: electron transfer flavoprotein alpha/beta-subunit;PDBTitle: electron transferring flavoprotein of acidaminococcus fermentans:2 towards a mechanism of flavin-based electron bifurcation
30 d1efvb_
not modelled
91.8
19
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits
31 d1wy5a1
not modelled
90.2
17
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: PP-loop ATPase
32 d1efpb_
not modelled
89.6
22
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits
33 c4l2iA_
not modelled
86.7
17
PDB header: electron transportChain: A: PDB Molecule: electron transfer flavoprotein alpha subunit;PDBTitle: electron transferring flavoprotein of acidaminococcus fermentans:2 towards a mechanism of flavin-based electron bifurcation
34 c5gafi_
not modelled
86.1
18
PDB header: ribosomeChain: I: PDB Molecule: 50s ribosomal protein l10;PDBTitle: rnc in complex with srp
35 c3zquA_
not modelled
80.3
8
PDB header: lyaseChain: A: PDB Molecule: probable aromatic acid decarboxylase;PDBTitle: structure of a probable aromatic acid decarboxylase
36 c3a2kB_
not modelled
77.9
17
PDB header: ligase/rnaChain: B: PDB Molecule: trna(ile)-lysidine synthase;PDBTitle: crystal structure of tils complexed with trna
37 c2j289_
not modelled
72.9
17
PDB header: ribosomeChain: 9: PDB Molecule: signal recognition particle 54;PDBTitle: model of e. coli srp bound to 70s rncs
38 c1o94D_
not modelled
72.4
12
PDB header: electron transportChain: D: PDB Molecule: electron transfer flavoprotein alpha-subunit;PDBTitle: ternary complex between trimethylamine dehydrogenase and2 electron transferring flavoprotein
39 c2e21A_
not modelled
68.8
17
PDB header: ligaseChain: A: PDB Molecule: trna(ile)-lysidine synthase;PDBTitle: crystal structure of tils in a complex with amppnp from aquifex2 aeolicus.
40 d2iela1
not modelled
65.5
13
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: Universal stress protein-like
41 c2iy3A_
not modelled
59.8
24
PDB header: rna-bindingChain: A: PDB Molecule: signal recognition particle protein,signal recognitionPDBTitle: structure of the e. coli signal regognition particle
42 c6jddA_
not modelled
58.2
14
PDB header: biosynthetic proteinChain: A: PDB Molecule: cypemycin cysteine dehydrogenase (decarboxylating);PDBTitle: crystal structure of the cypemycin decarboxylase cypd.
43 d1rvga_
not modelled
58.0
16
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class II FBP aldolase
44 c3ih5A_
not modelled
57.7
18
PDB header: electron transportChain: A: PDB Molecule: electron transfer flavoprotein alpha-subunit;PDBTitle: crystal structure of electron transfer flavoprotein alpha-subunit from2 bacteroides thetaiotaomicron
45 d1g5qa_
not modelled
57.5
9
Fold: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDSuperfamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDFamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
46 c4u7jB_
not modelled
56.3
16
PDB header: ligaseChain: B: PDB Molecule: argininosuccinate synthase;PDBTitle: crystal structure of argininosuccinate synthase from mycobacterium2 thermoresistibile
47 c6fahE_
not modelled
55.9
13
PDB header: flavoproteinChain: E: PDB Molecule: caffeyl-coa reductase-etf complex subunit care;PDBTitle: molecular basis of the flavin-based electron-bifurcating caffeyl-coa2 reductase reaction
48 c2j37W_
not modelled
54.5
16
PDB header: ribosomeChain: W: PDB Molecule: signal recognition particle 54 kda protein (srp54);PDBTitle: model of mammalian srp bound to 80s rncs
49 c2ejbA_
not modelled
51.4
6
PDB header: lyaseChain: A: PDB Molecule: probable aromatic acid decarboxylase;PDBTitle: crystal structure of phenylacrylic acid decarboxylase from2 aquifex aeolicus
50 d1p3y1_
not modelled
50.8
16
Fold: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDSuperfamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDFamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
51 c5udwB_
not modelled
49.9
18
PDB header: transferaseChain: B: PDB Molecule: lactate racemization operon protein lare;PDBTitle: lare, a sulfur transferase involved in synthesis of the cofactor for2 lactate racemase, in complex with nickel
52 c6eoaA_
not modelled
48.1
24
PDB header: flavoproteinChain: A: PDB Molecule: phosphopantothenoylcysteine decarboxylase;PDBTitle: crystal structure of hal3 from cryptococcus neoformans
53 d1qzua_
not modelled
47.9
17
Fold: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDSuperfamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDFamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
54 c3qjgD_
not modelled
46.9
12
PDB header: oxidoreductaseChain: D: PDB Molecule: epidermin biosynthesis protein epid;PDBTitle: epidermin biosynthesis protein epid from staphylococcus aureus
55 d3clsd1
not modelled
46.6
14
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits
56 c3dm5A_
not modelled
45.3
15
PDB header: rna binding protein, transport proteinChain: A: PDB Molecule: signal recognition 54 kda protein;PDBTitle: structures of srp54 and srp19, the two proteins assembling the2 ribonucleic core of the signal recognition particle from the archaeon3 pyrococcus furiosus.
57 c6qlgD_
not modelled
44.8
12
PDB header: transferaseChain: D: PDB Molecule: flavin prenyltransferase pad1, mitochondrial;PDBTitle: crystal structure of anubix (pada1) in complex with fmn and2 dimethylallyl pyrophosphate
58 c6ofuC_
not modelled
44.5
12
PDB header: lyaseChain: C: PDB Molecule: ydji aldolase;PDBTitle: x-ray crystal structure of the ydji aldolase from escherichia coli k12
59 c4nzpA_
not modelled
44.0
21
PDB header: ligaseChain: A: PDB Molecule: argininosuccinate synthase;PDBTitle: the crystal structure of argininosuccinate synthase from campylobacter2 jejuni subsp. jejuni nctc 11168
60 d1j20a1
not modelled
43.9
15
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: N-type ATP pyrophosphatases
61 d2dbsa1
not modelled
43.4
20
Fold: TTHC002-likeSuperfamily: TTHC002-likeFamily: TTHC002-like
62 c5h75B_
not modelled
42.8
18
PDB header: lyaseChain: B: PDB Molecule: mersacidin decarboxylase,immunoglobulin g-binding proteinPDBTitle: crystal structure of the mrsd-protein a fusion protein
63 c4rheB_
not modelled
41.4
15
PDB header: lyaseChain: B: PDB Molecule: 3-octaprenyl-4-hydroxybenzoate carboxy-lyase;PDBTitle: crystal structure of ubix, an aromatic acid decarboxylase from the2 colwellia psychrerythraea 34h
64 d1zuna1
not modelled
41.0
7
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: PAPS reductase-like
65 c2xdqB_
not modelled
40.3
25
PDB header: oxidoreductaseChain: B: PDB Molecule: light-independent protochlorophyllide reductase subunit b;PDBTitle: dark operative protochlorophyllide oxidoreductase (chln-chlb)2 complex
66 c3pm6B_
not modelled
39.9
14
PDB header: lyaseChain: B: PDB Molecule: putative fructose-bisphosphate aldolase;PDBTitle: crystal structure of a putative fructose-1,6-biphosphate aldolase from2 coccidioides immitis solved by combined sad mr
67 d1nu0a_
not modelled
39.6
18
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Putative Holliday junction resolvase RuvX
68 c3mcuF_
not modelled
38.6
16
PDB header: oxidoreductaseChain: F: PDB Molecule: dipicolinate synthase, b chain;PDBTitle: crystal structure of the dipicolinate synthase chain b from bacillus2 cereus. northeast structural genomics consortium target bcr215.
69 d1gvfa_
not modelled
37.4
10
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class II FBP aldolase
70 c1ni5A_
not modelled
35.8
19
PDB header: cell cycleChain: A: PDB Molecule: putative cell cycle protein mesj;PDBTitle: structure of the mesj pp-atpase from escherichia coli
71 c3clrD_
not modelled
34.3
12
PDB header: electron transportChain: D: PDB Molecule: electron transfer flavoprotein subunit alpha;PDBTitle: crystal structure of the r236a etf mutant from m. methylotrophus
72 c3pdiG_
not modelled
30.8
15
PDB header: protein bindingChain: G: PDB Molecule: nitrogenase mofe cofactor biosynthesis protein nife;PDBTitle: precursor bound nifen
73 c1zunA_
not modelled
30.6
7
PDB header: transferaseChain: A: PDB Molecule: sulfate adenylyltransferase subunit 2;PDBTitle: crystal structure of a gtp-regulated atp sulfurylase2 heterodimer from pseudomonas syringae
74 c2xdqA_
not modelled
30.4
6
PDB header: oxidoreductaseChain: A: PDB Molecule: light-independent protochlorophyllide reductase subunit n;PDBTitle: dark operative protochlorophyllide oxidoreductase (chln-chlb)2 complex
75 d1dd9a_
not modelled
30.0
13
Fold: DNA primase coreSuperfamily: DNA primase coreFamily: DNA primase DnaG catalytic core
76 c1dd9A_
not modelled
30.0
13
PDB header: transferaseChain: A: PDB Molecule: dna primase;PDBTitle: structure of the dnag catalytic core
77 c1k97A_
not modelled
29.9
21
PDB header: ligaseChain: A: PDB Molecule: argininosuccinate synthase;PDBTitle: crystal structure of e. coli argininosuccinate synthetase in complex2 with aspartate and citrulline
78 d1t6t1_
not modelled
29.8
11
Fold: Toprim domainSuperfamily: Toprim domainFamily: Toprim domain
79 d1sbza_
not modelled
29.1
11
Fold: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDSuperfamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDFamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
80 c6jlsA_
not modelled
28.6
23
PDB header: biosynthetic proteinChain: A: PDB Molecule: putative flavoprotein decarboxylase;PDBTitle: crystal structure of fmn-dependent cysteine decarboxylases tvaf from2 thioviridamide biosynthesis
81 c3vrhA_
not modelled
26.7
19
PDB header: rna binding proteinChain: A: PDB Molecule: putative uncharacterized protein ph0300;PDBTitle: crystal structure of ph0300
82 d1uyra2
not modelled
26.6
16
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Biotin dependent carboxylase carboxyltransferase domain
83 c3tvsA_
not modelled
26.5
11
PDB header: signaling proteinChain: A: PDB Molecule: cryptochrome-1;PDBTitle: structure of full-length drosophila cryptochrome
84 c4qskB_
not modelled
26.0
14
PDB header: ligaseChain: B: PDB Molecule: pyruvate carboxylase;PDBTitle: crystal structure of l. monocytogenes pyruvate carboxylase in complex2 with cyclic-di-amp
85 d1gph11
not modelled
24.3
12
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
86 d1gsoa2
not modelled
23.7
50
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like
87 d1vbka1
not modelled
23.2
11
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ThiI-like
88 c3iv7B_
not modelled
21.2
20
PDB header: oxidoreductaseChain: B: PDB Molecule: alcohol dehydrogenase iv;PDBTitle: crystal structure of iron-containing alcohol dehydrogenase2 (np_602249.1) from corynebacterium glutamicum atcc 13032 kitasato at3 2.07 a resolution
89 c2v3cC_
not modelled
21.1
23
PDB header: signaling proteinChain: C: PDB Molecule: signal recognition 54 kda protein;PDBTitle: crystal structure of the srp54-srp19-7s.s srp rna complex2 of m. jannaschii
90 c5l3rC_
not modelled
20.9
17
PDB header: protein transportChain: C: PDB Molecule: signal recognition particle 54 kda protein, chloroplastic;PDBTitle: structure of the gtpase heterodimer of chloroplast srp54 and ftsy from2 arabidopsis thaliana
91 d1ecfa1
not modelled
20.7
12
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
92 d1xkya1
not modelled
20.5
9
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
93 d1mvla_
not modelled
20.5
12
Fold: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDSuperfamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDFamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
94 c1mvlA_
not modelled
20.5
12
PDB header: lyaseChain: A: PDB Molecule: ppc decarboxylase athal3a;PDBTitle: ppc decarboxylase mutant c175s
95 c2j7pA_
not modelled
20.4
25
PDB header: signal recognitionChain: A: PDB Molecule: signal recognition particle protein;PDBTitle: gmppnp-stabilized ng domain complex of the srp gtpases ffh2 and ftsy
96 d1nuia1
not modelled
20.0
20
Fold: DNA primase coreSuperfamily: DNA primase coreFamily: Primase fragment of primase-helicase protein
97 d3bzka5
not modelled
20.0
21
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Tex RuvX-like domain-like
98 d1ru8a_
not modelled
19.9
18
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: N-type ATP pyrophosphatases
99 c5l3sF_
not modelled
19.8
12
PDB header: protein transportChain: F: PDB Molecule: signal recognition particle receptor ftsy;PDBTitle: structure of the gtpase heterodimer of crenarchaeal srp54 and ftsy