| 1 |
|
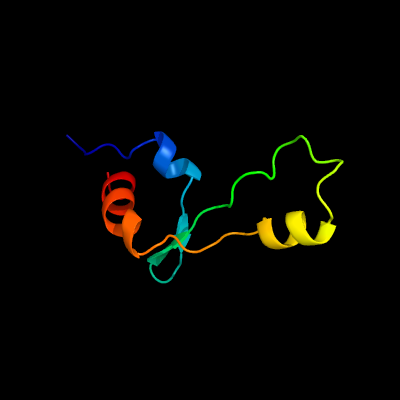
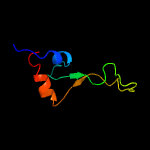
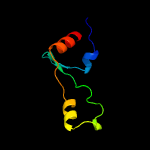
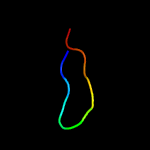
PDB 5o60 chain E
Region: 2 - 64
Aligned: 63
Modelled: 63
Confidence: 100.0%
Identity: 84%
PDB header:ribosome
Chain: E: PDB Molecule:50s ribosomal protein l4;
PDBTitle: structure of the 50s large ribosomal subunit from mycobacterium2 smegmatis
Phyre2
| 2 |
|
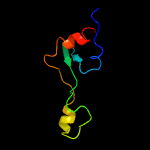
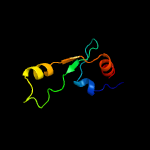
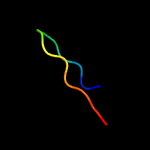
PDB 2qam chain 3 domain 1
Region: 2 - 63
Aligned: 62
Modelled: 62
Confidence: 100.0%
Identity: 40%
Fold: L35p-like
Superfamily: L35p-like
Family: Ribosomal protein L35p
Phyre2
| 3 |
|
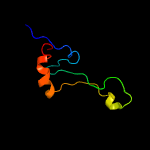
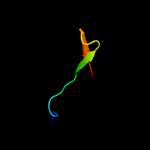
PDB 2j01 chain 8 domain 1
Region: 2 - 63
Aligned: 62
Modelled: 62
Confidence: 100.0%
Identity: 39%
Fold: L35p-like
Superfamily: L35p-like
Family: Ribosomal protein L35p
Phyre2
| 4 |
|
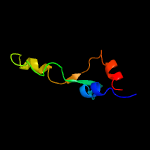
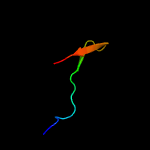
PDB 3bbo chain 5
Region: 3 - 63
Aligned: 61
Modelled: 61
Confidence: 100.0%
Identity: 44%
PDB header:ribosome
Chain: 5: PDB Molecule:ribosomal protein l35;
PDBTitle: homology model for the spinach chloroplast 50s subunit fitted to 9.4a2 cryo-em map of the 70s chlororibosome
Phyre2
| 5 |
|
PDB 2zjr chain 3 domain 1
Region: 2 - 63
Aligned: 62
Modelled: 62
Confidence: 100.0%
Identity: 34%
Fold: L35p-like
Superfamily: L35p-like
Family: Ribosomal protein L35p
Phyre2
| 6 |
|
PDB 4wfb chain 3
Region: 2 - 61
Aligned: 60
Modelled: 60
Confidence: 100.0%
Identity: 42%
PDB header:ribosome
Chain: 3: PDB Molecule:50s ribosomal protein l35;
PDBTitle: the crystal structure of the large ribosomal subunit of staphylococcus2 aureus in complex with bc-3205
Phyre2
| 7 |
|
PDB 1vw4 chain Z
Region: 4 - 63
Aligned: 59
Modelled: 60
Confidence: 99.6%
Identity: 34%
PDB header:ribosome
Chain: Z: PDB Molecule:mitochondrial ribosomal protein ynl122c;
PDBTitle: structure of the yeast mitochondrial large ribosomal subunit
Phyre2
| 8 |
|
PDB 4v19 chain 8
Region: 3 - 62
Aligned: 60
Modelled: 60
Confidence: 97.6%
Identity: 28%
PDB header:ribosome
Chain: 8: PDB Molecule:mitoribosomal protein bl35m, mrpl35;
PDBTitle: structure of the large subunit of the mammalian mitoribosome, part 12 of 2
Phyre2
| 9 |
|
PDB 4ce4 chain 8
Region: 3 - 61
Aligned: 59
Modelled: 59
Confidence: 97.3%
Identity: 29%
PDB header:ribosome
Chain: 8: PDB Molecule:mrpl35;
PDBTitle: 39s large subunit of the porcine mitochondrial ribosome
Phyre2
| 10 |
|
PDB 4byl chain 5
Region: 14 - 26
Aligned: 13
Modelled: 13
Confidence: 9.1%
Identity: 15%
PDB header:ribosome
Chain: 5: PDB Molecule:ubiquitin-40s ribosomal protein s31;
PDBTitle: cryo-em reconstruction of the 80s-eif5b-met-itrnamet2 eukaryotic translation initiation complex
Phyre2
| 11 |
|
PDB 3u5c chain F
Region: 14 - 26
Aligned: 13
Modelled: 13
Confidence: 9.1%
Identity: 15%
PDB header:ribosome
Chain: F: PDB Molecule:40s ribosomal protein s5;
PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution. this2 entry contains proteins of the 40s subunit, ribosome a
Phyre2
| 12 |
|
PDB 4byt chain 5
Region: 14 - 26
Aligned: 13
Modelled: 13
Confidence: 9.1%
Identity: 15%
PDB header:ribosome
Chain: 5: PDB Molecule:ubiquitin-40s ribosomal protein s31;
PDBTitle: cryo-em reconstruction of the 80s-eif5b-met-itrnamet2 eukaryotic translation initiation complex
Phyre2
| 13 |
|
PDB 4uer chain 9
Region: 14 - 26
Aligned: 13
Modelled: 13
Confidence: 9.1%
Identity: 15%
PDB header:translation
Chain: 9: PDB Molecule:es31;
PDBTitle: 40s-eif1-eif1a-eif3-eif3j translation initiation complex from2 lachancea kluyveri
Phyre2
| 14 |
|
PDB 2m7e chain A
Region: 11 - 19
Aligned: 9
Modelled: 9
Confidence: 8.0%
Identity: 44%
PDB header:calmodulin-binding protein
Chain: A: PDB Molecule:calcium-transporting atpase 2, plasma membrane-type;
PDBTitle: solution structure of the calmodulin-binding domain of plant calcium-2 atpase aca2
Phyre2
| 15 |
|
PDB 5xxu chain F
Region: 14 - 26
Aligned: 13
Modelled: 13
Confidence: 7.8%
Identity: 23%
PDB header:ribosome
Chain: F: PDB Molecule:ribosomal protein us7;
PDBTitle: small subunit of toxoplasma gondii ribosome
Phyre2
| 16 |
|
PDB 2xzm chain 9
Region: 5 - 26
Aligned: 22
Modelled: 22
Confidence: 6.5%
Identity: 27%
PDB header:ribosome
Chain: 9: PDB Molecule:rps31e;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
Phyre2
| 17 |
|
PDB 2xzn chain 9
Region: 5 - 26
Aligned: 22
Modelled: 22
Confidence: 6.5%
Identity: 27%
PDB header:ribosome
Chain: 9: PDB Molecule:rps31e;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
Phyre2
| 18 |
|
PDB 2l53 chain B
Region: 25 - 33
Aligned: 9
Modelled: 9
Confidence: 6.3%
Identity: 78%
PDB header:ca-binding protein/proton transport
Chain: B: PDB Molecule:voltage-gated sodium channel type v alpha isoform b
PDBTitle: solution nmr structure of apo-calmodulin in complex with the iq motif2 of human cardiac sodium channel nav1.5
Phyre2
| 19 |
|
PDB 3j38 chain F
Region: 5 - 26
Aligned: 22
Modelled: 22
Confidence: 5.6%
Identity: 27%
PDB header:ribosome
Chain: F: PDB Molecule:40s ribosomal protein s5a;
PDBTitle: structure of the d. melanogaster 40s ribosomal proteins
Phyre2