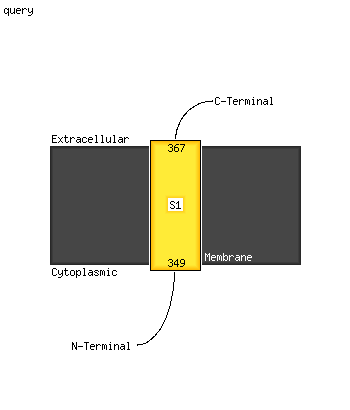
| 1 | c5mu5A_
|
|
 |
99.5 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: structure of maf glycosyltransferase from magnetospirillum magneticum2 amb-1
|
|
|
|
| 2 | c3lm8D_
|
|
 |
99.3 |
25 |
PDB header:transferase
Chain: D: PDB Molecule:thiamine pyrophosphokinase;
PDBTitle: crystal structure of thiamine pyrophosphokinase from bacillus2 subtilis, northeast structural genomics consortium target sr677
|
|
|
|
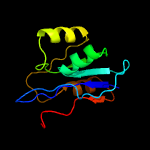
| 3 | c3ihkC_
|
|
 |
99.2 |
27 |
PDB header:transferase
Chain: C: PDB Molecule:thiamin pyrophosphokinase;
PDBTitle: crystal structure of thiamin pyrophosphokinase from s.mutans,2 northeast structural genomics consortium target smr83
|
|
|
|
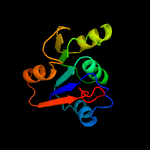
| 4 | c3melC_
|
|
 |
99.1 |
26 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:thiamin pyrophosphokinase family protein;
PDBTitle: crystal structure of thiamin pyrophosphokinase family protein from2 enterococcus faecalis, northeast structural genomics consortium3 target efr150
|
|
|
|
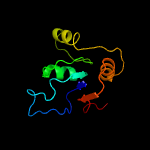
| 5 | c3l8mA_
|
|
 |
99.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:probable thiamine pyrophosphokinase;
PDBTitle: crystal structure of a probable thiamine pyrophosphokinase2 from staphylococcus saprophyticus subsp. saprophyticus.3 northeast structural genomics consortium target id syr86
|
|
|
|
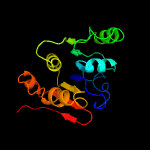
| 6 | c3cq9C_
|
|
 |
99.0 |
22 |
PDB header:transferase
Chain: C: PDB Molecule:uncharacterized protein lp_1622;
PDBTitle: crystal structure of the lp_1622 protein from lactobacillus plantarum.2 northeast structural genomics consortium target lpr114
|
|
|
|
| 7 | c3k94A_
|
|
 |
99.0 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:thiamin pyrophosphokinase;
PDBTitle: crystal structure of thiamin pyrophosphokinase from geobacillus2 thermodenitrificans, northeast structural genomics consortium target3 gtr2
|
|
|
|
| 8 | c2f17A_
|
|
 |
98.7 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:thiamin pyrophosphokinase 1;
PDBTitle: mouse thiamin pyrophosphokinase in a ternary complex with2 pyrithiamin pyrophosphate and amp at 2.5 angstrom
|
|
|
|
| 9 | c1ig0A_
|
|
 |
98.7 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:thiamin pyrophosphokinase;
PDBTitle: crystal structure of yeast thiamin pyrophosphokinase
|
|
|
|
| 10 | c2hh9A_
|
|
 |
98.7 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:thiamin pyrophosphokinase;
PDBTitle: thiamin pyrophosphokinase from candida albicans
|
|
|
|
| 11 | d1ig0a2
|
|
 |
98.6 |
22 |
Fold:Thiamin pyrophosphokinase, catalytic domain
Superfamily:Thiamin pyrophosphokinase, catalytic domain
Family:Thiamin pyrophosphokinase, catalytic domain |
|
|
|
| 12 | d1ig3a2
|
|
 |
98.6 |
23 |
Fold:Thiamin pyrophosphokinase, catalytic domain
Superfamily:Thiamin pyrophosphokinase, catalytic domain
Family:Thiamin pyrophosphokinase, catalytic domain |
|
|
|
| 13 | c2omkB_
|
|
 |
98.3 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:hypothetical protein;
PDBTitle: structure of the bacteroides thetaiotaomicron thiamin2 pyrophosphokinase
|
|
|
|
| 14 | c2e28A_
|
|
 |
95.6 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure analysis of pyruvate kinase from bacillus2 stearothermophilus
|
|
|
|
| 15 | c4j0nA_
|
|
 |
93.3 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:isatin hydrolase b;
PDBTitle: crystal structure of a manganese dependent isatin hydrolase
|
|
|
|
| 16 | c5ibzD_
|
|
 |
93.1 |
30 |
PDB header:lyase
Chain: D: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a novel cyclase (pfam04199).
|
|
|
|
| 17 | c5fbtA_
|
|
 |
92.7 |
21 |
PDB header:transferase/antibiotic
Chain: A: PDB Molecule:phosphoenolpyruvate synthase;
PDBTitle: crystal structure of rifampin phosphotransferase rph-lm from listeria2 monocytogenes in complex with rifampin
|
|
|
|
| 18 | d1kbla2
|
|
 |
92.4 |
18 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Phosphohistidine domain
Family:Pyruvate phosphate dikinase, central domain |
|
|
|
| 19 | d1ytla1
|
|
 |
91.0 |
17 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:ACDE2-like |
|
|
|
| 20 | d1zyma2
|
|
 |
90.4 |
17 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Phosphohistidine domain
Family:N-terminal domain of enzyme I of the PEP:sugar phosphotransferase system |
|
|
|
| 21 | d1vbga2 |
|
not modelled |
90.4 |
13 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Phosphohistidine domain
Family:Pyruvate phosphate dikinase, central domain |
|
|
| 22 | c3t07D_ |
|
not modelled |
90.3 |
12 |
PDB header:transferase/transferase inhibitor
Chain: D: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of s. aureus pyruvate kinase in complex with a2 naturally occurring bis-indole alkaloid
|
|
|
| 23 | c5nnaB_ |
|
not modelled |
89.6 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:isatin hydrolase a;
PDBTitle: isatin hydrolase a (iha) from labrenzia aggregata bound to benzyl2 benzoate
|
|
|
| 24 | d1h6za2 |
|
not modelled |
89.4 |
16 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Phosphohistidine domain
Family:Pyruvate phosphate dikinase, central domain |
|
|
| 25 | c4coaA_ |
|
not modelled |
87.2 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:kynurenine formamidase;
PDBTitle: crystal structure of kynurenine formamidase from bacillus2 anthracis complexed with 2-aminoacetophenone.
|
|
|
| 26 | d1r61a_ |
|
not modelled |
86.1 |
22 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Putative cyclase
Family:Putative cyclase |
|
|
| 27 | c6ar9A_ |
|
not modelled |
83.5 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:hypoxanthine-guanine phosphoribosyltransferase, putative;
PDBTitle: crystal structure of hypoxanthine-guanine-xanthine2 phosphorybosyltranferase in complex with [(2-{[2-(2-amino-6-oxo-1,6-3 dihydro-9h-purin-9-yl)ethyl][(e)-2-phosphonoethenyl]amino}ethoxy)4 methyl]phosphonic acid
|
|
|
| 28 | c2vt2A_ |
|
not modelled |
83.1 |
24 |
PDB header:transcription
Chain: A: PDB Molecule:redox-sensing transcriptional repressor rex;
PDBTitle: structure and functional properties of the bacillus subtilis2 transcriptional repressor rex
|
|
|
| 29 | c4cobA_ |
|
not modelled |
82.9 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:kynurenine formamidase;
PDBTitle: crystal structure kynurenine formamidase from pseudomonas aeruginosa
|
|
|
| 30 | d2hi6a1 |
|
not modelled |
82.2 |
28 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:LeuD/IlvD-like
Family:AF0055-like |
|
|
| 31 | c4cogB_ |
|
not modelled |
81.7 |
10 |
PDB header:hydrolase
Chain: B: PDB Molecule:kynurenine formamidase;
PDBTitle: crystal structure of kynurenine formamidase from burkholderia2 cenocepacia
|
|
|
| 32 | c5nmpF_ |
|
not modelled |
80.0 |
15 |
PDB header:hydrolase
Chain: F: PDB Molecule:isatin hydrolase;
PDBTitle: isatin hydrolase a (iha) from ralstonia solanacearum
|
|
|
| 33 | d1pjqa1 |
|
not modelled |
78.9 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Siroheme synthase N-terminal domain-like |
|
|
| 34 | d2b0aa1 |
|
not modelled |
78.1 |
17 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Putative cyclase
Family:Putative cyclase |
|
|
| 35 | c5l9wB_ |
|
not modelled |
78.1 |
20 |
PDB header:ligase
Chain: B: PDB Molecule:acetophenone carboxylase gamma subunit;
PDBTitle: crystal structure of the apc core complex
|
|
|
| 36 | c4cjxA_ |
|
not modelled |
76.2 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:c-1-tetrahydrofolate synthase, cytoplasmic, putative;
PDBTitle: the crystal structure of trypanosoma brucei n5, n10-2 methylenetetrahydrofolate dehydrogenase-cyclohydrolase (fold)3 complexed with nadp cofactor and inhibitor
|
|
|
| 37 | d1kyqa1 |
|
not modelled |
76.1 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Siroheme synthase N-terminal domain-like |
|
|
| 38 | c4ldaF_ |
|
not modelled |
75.8 |
16 |
PDB header:transcription
Chain: F: PDB Molecule:tadz;
PDBTitle: crystal structure of a chey-like protein (tadz) from pseudomonas2 aeruginosa pao1 at 2.70 a resolution
|
|
|
| 39 | c4b4uB_ |
|
not modelled |
75.2 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of acinetobacter baumannii n5,2 n10-methylenetetrahydrofolate dehydrogenase-cyclohydrolase (fold)3 complexed with nadp cofactor
|
|
|
| 40 | d2jfga1 |
|
not modelled |
74.8 |
14 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
|
|
| 41 | c1kyqC_ |
|
not modelled |
74.7 |
15 |
PDB header:oxidoreductase, lyase
Chain: C: PDB Molecule:siroheme biosynthesis protein met8;
PDBTitle: met8p: a bifunctional nad-dependent dehydrogenase and2 ferrochelatase involved in siroheme synthesis.
|
|
|
| 42 | c3l07B_ |
|
not modelled |
73.5 |
19 |
PDB header:oxidoreductase,hydrolase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate2 cyclohydrolase, putative bifunctional protein fold from francisella3 tularensis.
|
|
|
| 43 | c5xoeA_ |
|
not modelled |
70.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:atp-dependent 6-phosphofructokinase;
PDBTitle: crystal structure of the apo staphylococcus aureus phosphofructokinase
|
|
|
| 44 | c2c2xB_ |
|
not modelled |
69.9 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:methylenetetrahydrofolate dehydrogenase-
PDBTitle: three dimensional structure of bifunctional methylenetetrahydrofolate2 dehydrogenase-cyclohydrolase from mycobacterium tuberculosis
|
|
|
| 45 | c3nglA_ |
|
not modelled |
69.6 |
19 |
PDB header:oxidoreductase, hydrolase
Chain: A: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of bifunctional 5,10-methylenetetrahydrofolate2 dehydrogenase / cyclohydrolase from thermoplasma acidophilum
|
|
|
| 46 | c3opyG_ |
|
not modelled |
69.4 |
19 |
PDB header:transferase
Chain: G: PDB Molecule:6-phosphofructo-1-kinase alpha-subunit;
PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
|
|
|
| 47 | c1a4iB_ |
|
not modelled |
69.1 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:methylenetetrahydrofolate dehydrogenase /
PDBTitle: human tetrahydrofolate dehydrogenase / cyclohydrolase
|
|
|
| 48 | c3o8nA_ |
|
not modelled |
68.8 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:6-phosphofructokinase, muscle type;
PDBTitle: structure of phosphofructokinase from rabbit skeletal muscle
|
|
|
| 49 | c5zf1A_ |
|
not modelled |
68.6 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:5,10-methylenetetrahydrofolate dehydrogenase;
PDBTitle: molecular structure of a novel 5,10-methylenetetrahydrofolate2 dehydrogenase from the silkworm, bombyx mori
|
|
|
| 50 | c1edzA_ |
|
not modelled |
68.3 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:5,10-methylenetetrahydrofolate dehydrogenase;
PDBTitle: structure of the nad-dependent 5,10-2 methylenetetrahydrofolate dehydrogenase from saccharomyces3 cerevisiae
|
|
|
| 51 | d1b0aa1 |
|
not modelled |
66.8 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
|
|
| 52 | c3p2oA_ |
|
not modelled |
66.2 |
19 |
PDB header:oxidoreductase, hydrolase
Chain: A: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of fold bifunctional protein from campylobacter2 jejuni
|
|
|
| 53 | c3opyE_ |
|
not modelled |
65.9 |
19 |
PDB header:transferase
Chain: E: PDB Molecule:6-phosphofructo-1-kinase alpha-subunit;
PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
|
|
|
| 54 | c2dt5A_ |
|
not modelled |
65.3 |
31 |
PDB header:dna binding protein
Chain: A: PDB Molecule:at-rich dna-binding protein;
PDBTitle: crystal structure of ttha1657 (at-rich dna-binding protein) from2 thermus thermophilus hb8
|
|
|
| 55 | c4ehiB_ |
|
not modelled |
64.7 |
14 |
PDB header:hydrolase,transferase
Chain: B: PDB Molecule:bifunctional purine biosynthesis protein purh;
PDBTitle: an x-ray crystal structure of a putative bifunctional2 phosphoribosylaminoimidazolecarboxamide formyltransferase/imp3 cyclohydrolase
|
|
|
| 56 | c5x5jA_ |
|
not modelled |
64.5 |
13 |
PDB header:dna binding protein
Chain: A: PDB Molecule:ader;
PDBTitle: crystal structure of response regulator ader receiver domain
|
|
|
| 57 | c6nkoA_ |
|
not modelled |
64.3 |
18 |
PDB header:unknown function
Chain: A: PDB Molecule:forh;
PDBTitle: crystal structure of forh
|
|
|
| 58 | d4pfka_ |
|
not modelled |
63.4 |
20 |
Fold:Phosphofructokinase
Superfamily:Phosphofructokinase
Family:Phosphofructokinase |
|
|
| 59 | c6apeA_ |
|
not modelled |
62.8 |
23 |
PDB header:oxidoreductase, hydrolase
Chain: A: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of bifunctional protein fold from helicobacter2 pylori
|
|
|
| 60 | c3opyB_ |
|
not modelled |
62.5 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:6-phosphofructo-1-kinase beta-subunit;
PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
|
|
|
| 61 | c3opyH_ |
|
not modelled |
62.5 |
11 |
PDB header:transferase
Chain: H: PDB Molecule:6-phosphofructo-1-kinase beta-subunit;
PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
|
|
|
| 62 | c6d73C_ |
|
not modelled |
61.9 |
31 |
PDB header:transport protein
Chain: C: PDB Molecule:transient receptor potential cation channel, subfamily m;
PDBTitle: cryo-em structure of the zebrafish trpm2 channel in the presence of2 ca2+
|
|
|
| 63 | c3zpgA_ |
|
not modelled |
61.4 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:ribd;
PDBTitle: acinetobacter baumannii ribd, form 2
|
|
|
| 64 | d1a4ia1 |
|
not modelled |
61.3 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
|
|
| 65 | c4a5oB_ |
|
not modelled |
61.0 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of pseudomonas aeruginosa n5, n10-2 methylenetetrahydrofolate dehydrogenase-cyclohydrolase (fold)
|
|
|
| 66 | c6bpqC_ |
|
not modelled |
60.7 |
32 |
PDB header:transport protein
Chain: C: PDB Molecule:transient receptor potential cation channel subfamily m
PDBTitle: structure of the cold- and menthol-sensing ion channel trpm8
|
|
|
| 67 | c6bpqD_ |
|
not modelled |
60.7 |
32 |
PDB header:transport protein
Chain: D: PDB Molecule:transient receptor potential cation channel subfamily m
PDBTitle: structure of the cold- and menthol-sensing ion channel trpm8
|
|
|
| 68 | c6bpqA_ |
|
not modelled |
60.7 |
32 |
PDB header:transport protein
Chain: A: PDB Molecule:transient receptor potential cation channel subfamily m
PDBTitle: structure of the cold- and menthol-sensing ion channel trpm8
|
|
|
| 69 | c6bpqB_ |
|
not modelled |
60.7 |
32 |
PDB header:transport protein
Chain: B: PDB Molecule:transient receptor potential cation channel subfamily m
PDBTitle: structure of the cold- and menthol-sensing ion channel trpm8
|
|
|
| 70 | c3p2oB_ |
|
not modelled |
60.1 |
17 |
PDB header:oxidoreductase, hydrolase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of fold bifunctional protein from campylobacter2 jejuni
|
|
|
| 71 | c4a26B_ |
|
not modelled |
59.4 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative c-1-tetrahydrofolate synthase, cytoplasmic;
PDBTitle: the crystal structure of leishmania major n5,n10-2 methylenetetrahydrofolate dehydrogenase/cyclohydrolase
|
|
|
| 72 | c5nhsB_ |
|
not modelled |
59.2 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: the crystal structure of xanthomonas albilineans n5, n10-2 methylenetetrahydrofolate dehydrogenase-cyclohydrolase (fold)
|
|
|
| 73 | d1a9xa4 |
|
not modelled |
58.8 |
18 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
|
|
| 74 | d1lssa_ |
|
not modelled |
58.8 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Potassium channel NAD-binding domain |
|
|
| 75 | d1dbwa_ |
|
not modelled |
58.7 |
19 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 76 | c3nzoB_ |
|
not modelled |
58.3 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:udp-n-acetylglucosamine 4,6-dehydratase;
PDBTitle: udp-n-acetylglucosamine 4,6-dehydratase from vibrio fischeri.
|
|
|
| 77 | d1pfka_ |
|
not modelled |
57.4 |
18 |
Fold:Phosphofructokinase
Superfamily:Phosphofructokinase
Family:Phosphofructokinase |
|
|
| 78 | d2gv8a2 |
|
not modelled |
57.3 |
24 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
|
|
| 79 | c2yvqA_ |
|
not modelled |
56.8 |
11 |
PDB header:ligase
Chain: A: PDB Molecule:carbamoyl-phosphate synthase;
PDBTitle: crystal structure of mgs domain of carbamoyl-phosphate2 synthetase from homo sapiens
|
|
|
| 80 | c5zz5D_ |
|
not modelled |
55.9 |
26 |
PDB header:gene regulation
Chain: D: PDB Molecule:redox-sensing transcriptional repressor rex;
PDBTitle: redox-sensing transcriptional repressor rex
|
|
|
| 81 | d1mvoa_ |
|
not modelled |
55.6 |
14 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 82 | c1b0aA_ |
|
not modelled |
55.0 |
20 |
PDB header:oxidoreductase,hydrolase
Chain: A: PDB Molecule:protein (fold bifunctional protein);
PDBTitle: 5,10, methylene-tetrahydropholate2 dehydrogenase/cyclohydrolase from e coli.
|
|
|
| 83 | c5kojD_ |
|
not modelled |
54.6 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:nitrogenase femo beta subunit protein nifk;
PDBTitle: nitrogenase mofep protein in the ids oxidized state
|
|
|
| 84 | c5l9wb_ |
|
not modelled |
54.3 |
13 |
PDB header:ligase
Chain: B: PDB Molecule:acetophenone carboxylase gamma subunit;
PDBTitle: crystal structure of the apc core complex
|
|
|
| 85 | c3dfzB_ |
|
not modelled |
54.3 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:precorrin-2 dehydrogenase;
PDBTitle: sirc, precorrin-2 dehydrogenase
|
|
|
| 86 | d1v6sa_ |
|
not modelled |
54.2 |
21 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
|
|
| 87 | c1fcdB_ |
|
not modelled |
53.9 |
24 |
PDB header:electron transport(flavocytochrome)
Chain: B: PDB Molecule:flavocytochrome c sulfide dehydrogenase (flavin-
PDBTitle: the structure of flavocytochrome c sulfide dehydrogenase2 from a purple phototrophic bacterium chromatium vinosum at3 2.5 angstroms resolution
|
|
|
| 88 | c4plpB_ |
|
not modelled |
53.6 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:homospermidine synthase;
PDBTitle: crystal structure of the homospermidine synthase (hss) from2 blastochloris viridis in complex with nad
|
|
|
| 89 | c3snkA_ |
|
not modelled |
52.9 |
12 |
PDB header:signaling protein
Chain: A: PDB Molecule:response regulator chey-like protein;
PDBTitle: crystal structure of a response regulator chey-like protein (mll6475)2 from mesorhizobium loti at 2.02 a resolution
|
|
|
| 90 | d1fcda1 |
|
not modelled |
52.6 |
24 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
|
|
| 91 | c1pjtB_ |
|
not modelled |
51.2 |
16 |
PDB header:transferase/oxidoreductase/lyase
Chain: B: PDB Molecule:siroheme synthase;
PDBTitle: the structure of the ser128ala point-mutant variant of cysg, the2 multifunctional methyltransferase/dehydrogenase/ferrochelatase for3 siroheme synthesis
|
|
|
| 92 | d1dcfa_ |
|
not modelled |
51.0 |
9 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:Receiver domain of the ethylene receptor |
|
|
| 93 | c3trjC_ |
|
not modelled |
50.9 |
13 |
PDB header:isomerase
Chain: C: PDB Molecule:phosphoheptose isomerase;
PDBTitle: structure of a phosphoheptose isomerase from francisella tularensis
|
|
|
| 94 | c6od1A_ |
|
not modelled |
50.6 |
19 |
PDB header:signaling protein
Chain: A: PDB Molecule:regulator of rpos;
PDBTitle: irad-bound to rssb d58p variant
|
|
|
| 95 | c3vbaE_ |
|
not modelled |
50.2 |
22 |
PDB header:lyase
Chain: E: PDB Molecule:isopropylmalate/citramalate isomerase small subunit;
PDBTitle: crystal structure of methanogen 3-isopropylmalate isomerase small2 subunit
|
|
|
| 96 | c5m45K_ |
|
not modelled |
48.7 |
14 |
PDB header:ligase
Chain: K: PDB Molecule:acetone carboxylase beta subunit;
PDBTitle: structure of acetone carboxylase purified from xanthobacter2 autotrophicus
|
|
|
| 97 | d1cjba_ |
|
not modelled |
48.7 |
17 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
|
|
| 98 | c5u8mA_ |
|
not modelled |
48.6 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:response regulator;
PDBTitle: a novel family of redox sensors in the streptococci evolved from two-2 component response regulators
|
|
|
| 99 | d2f48a1 |
|
not modelled |
48.0 |
15 |
Fold:Phosphofructokinase
Superfamily:Phosphofructokinase
Family:Phosphofructokinase |
|
|
| 100 | c2xdqA_ |
|
not modelled |
47.6 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:light-independent protochlorophyllide reductase subunit n;
PDBTitle: dark operative protochlorophyllide oxidoreductase (chln-chlb)2 complex
|
|
|
| 101 | d3bzka5 |
|
not modelled |
47.3 |
23 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Tex RuvX-like domain-like |
|
|
| 102 | c4a1oB_ |
|
not modelled |
47.2 |
19 |
PDB header:transferase-hydrolase
Chain: B: PDB Molecule:bifunctional purine biosynthesis protein purh;
PDBTitle: crystal structure of mycobacterium tuberculosis purh complexed with2 aicar and a novel nucleotide cfair, at 2.48 a resolution.
|
|
|
| 103 | c4lzlA_ |
|
not modelled |
46.9 |
12 |
PDB header:transcription
Chain: A: PDB Molecule:response regulator;
PDBTitle: structure of the inactive form of the regulatory domain from the2 repressor of iron transport regulator (ritr)
|
|
|
| 104 | d1u0sy_ |
|
not modelled |
46.6 |
15 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 105 | c3wg9D_ |
|
not modelled |
46.6 |
31 |
PDB header:transcription
Chain: D: PDB Molecule:redox-sensing transcriptional repressor rex;
PDBTitle: crystal structure of rsp, a rex-family repressor
|
|
|
| 106 | c2dwcB_ |
|
not modelled |
45.9 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:433aa long hypothetical phosphoribosylglycinamide formyl
PDBTitle: crystal structure of probable phosphoribosylglycinamide formyl2 transferase from pyrococcus horikoshii ot3 complexed with adp
|
|
|
| 107 | d1fsga_ |
|
not modelled |
45.5 |
21 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
|
|
| 108 | c4dadA_ |
|
not modelled |
44.6 |
16 |
PDB header:signaling protein, signal transduction
Chain: A: PDB Molecule:putative pilus assembly-related protein;
PDBTitle: crystal structure of a putative pilus assembly-related protein2 (bpss2195) from burkholderia pseudomallei k96243 at 2.50 a resolution3 (psi community target, shapiro l.)
|
|
|
| 109 | c2higA_ |
|
not modelled |
44.3 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:6-phospho-1-fructokinase;
PDBTitle: crystal structure of phosphofructokinase apoenzyme from trypanosoma2 brucei.
|
|
|
| 110 | c3elbA_ |
|
not modelled |
44.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:ethanolamine-phosphate cytidylyltransferase;
PDBTitle: human ctp: phosphoethanolamine cytidylyltransferase in complex with2 cmp
|
|
|
| 111 | d1zcza1 |
|
not modelled |
43.6 |
15 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Inosicase |
|
|
| 112 | c3ketA_ |
|
not modelled |
43.4 |
27 |
PDB header:transcription/dna
Chain: A: PDB Molecule:redox-sensing transcriptional repressor rex;
PDBTitle: crystal structure of a rex-family transcriptional regulatory protein2 from streptococcus agalactiae bound to a palindromic operator
|
|
|
| 113 | d1fw8a_ |
|
not modelled |
43.2 |
13 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
|
|
| 114 | c1thzA_ |
|
not modelled |
43.1 |
20 |
PDB header:transferase, hydrolase
Chain: A: PDB Molecule:bifunctional purine biosynthesis protein purh;
PDBTitle: crystal structure of avian aicar transformylase in complex2 with a novel inhibitor identified by virtual ligand3 screening
|
|
|
| 115 | c2r8zC_ |
|
not modelled |
42.7 |
9 |
PDB header:hydrolase
Chain: C: PDB Molecule:3-deoxy-d-manno-octulosonate 8-phosphate phosphatase;
PDBTitle: crystal structure of yrbi phosphatase from escherichia coli in complex2 with a phosphate and a calcium ion
|
|
|
| 116 | d1jb9a1 |
|
not modelled |
42.1 |
25 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
|
|
| 117 | d1g8ma1 |
|
not modelled |
41.1 |
20 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Inosicase |
|
|
| 118 | d2b3za2 |
|
not modelled |
41.0 |
17 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Deoxycytidylate deaminase-like |
|
|
| 119 | c4tlmC_ |
|
not modelled |
40.2 |
13 |
PDB header:signaling protein
Chain: C: PDB Molecule:receptor subunit glun1;
PDBTitle: crystal structure of glun1/glun2b nmda receptor, structure 2
|
|
|
| 120 | c3g8qA_ |
|
not modelled |
40.2 |
43 |
PDB header:rna binding protein
Chain: A: PDB Molecule:predicted rna-binding protein, contains thump
PDBTitle: a cytidine deaminase edits c-to-u in transfer rnas in2 archaea
|
|
|