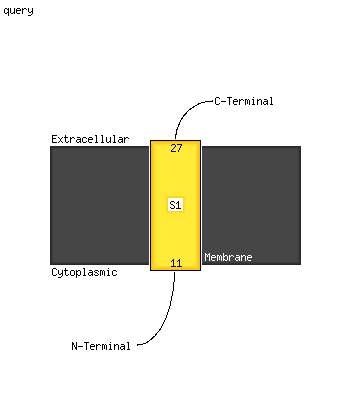
1 c1sy7B_
92.9
9
PDB header: oxidoreductaseChain: B: PDB Molecule: catalase 1;PDBTitle: crystal structure of the catalase-1 from neurospora crassa, native2 structure at 1.75a resolution.
2 d1p80a1
88.9
21
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Catalase, C-terminal domain
3 c1p81A_
88.3
19
PDB header: oxidoreductaseChain: A: PDB Molecule: catalase hpii;PDBTitle: crystal structure of the d181e variant of catalase hpii2 from e. coli
4 d1sy7a1
86.9
9
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Catalase, C-terminal domain
5 d1o57a2
82.0
15
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
6 d1mzva_
81.2
21
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
7 d1vcha1
80.4
31
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
8 c1o57A_
79.6
15
PDB header: dna binding proteinChain: A: PDB Molecule: pur operon repressor;PDBTitle: crystal structure of the purine operon repressor of2 bacillus subtilis
9 c4lzaB_
79.5
36
PDB header: transferaseChain: B: PDB Molecule: adenine phosphoribosyltransferase;PDBTitle: crystal structure of adenine phosphoribosyltransferase from2 thermoanaerobacter pseudethanolicus atcc 33223, nysgrc target 029700.
10 d1y0ba1
78.5
20
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
11 c2dy0A_
77.7
21
PDB header: transferaseChain: A: PDB Molecule: adenine phosphoribosyltransferase;PDBTitle: crystal structure of project jw0458 from escherichia coli
12 c3ej6D_
77.6
19
PDB header: oxidoreductaseChain: D: PDB Molecule: catalase-3;PDBTitle: neurospora crassa catalase-3 crystal structure
13 d1g2qa_
77.3
19
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
14 c4m0kD_
76.8
26
PDB header: transferaseChain: D: PDB Molecule: adenine phosphoribosyltransferase;PDBTitle: crystal structure of adenine phosphoribosyltransferase from2 rhodothermus marinus dsm 4252, nysgrc target 029775.
15 c2vrnA_
76.3
21
PDB header: hydrolaseChain: A: PDB Molecule: protease i;PDBTitle: the structure of the stress response protein dr1199 from deinococcus2 radiodurans: a member of the dj-1 superfamily
16 c5zgoB_
75.4
24
PDB header: transferaseChain: B: PDB Molecule: adenine phosphoribosyltransferase;PDBTitle: crystal structure of aprt2 from thermus thermophilus hb8
17 d1zn7a1
73.0
24
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
18 c5fv8B_
72.7
23
PDB header: structural proteinChain: B: PDB Molecule: fosw;PDBTitle: structure of cjun-fosw coiled coil complex.
19 c5fv8A_
72.7
23
PDB header: structural proteinChain: A: PDB Molecule: fosw;PDBTitle: structure of cjun-fosw coiled coil complex.
20 d1lh0a_
71.5
9
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
21 d1vmea1
not modelled
68.4
10
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related
22 c2wnsB_
not modelled
66.9
14
PDB header: transferaseChain: B: PDB Molecule: orotate phosphoribosyltransferase;PDBTitle: human orotate phosphoribosyltransferase (oprtase) domain of2 uridine 5'-monophosphate synthase (umps) in complex with3 its substrate orotidine 5'-monophosphate (omp)
23 c5hkiD_
not modelled
64.2
14
PDB header: transferaseChain: D: PDB Molecule: orotate phosphoribosyltransferase;PDBTitle: crystal structure of mycobacterium tuberculosis h37rv orotate2 phosphoribosyltransferase in complex with fe(iii) dicitrate
24 d2fz5a1
not modelled
64.1
17
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related
25 c5yw2D_
not modelled
60.7
12
PDB header: transferaseChain: D: PDB Molecule: adenine phosphoribosyltransferase;PDBTitle: crystal structure of adenine phosphoribosyltransferase from2 francisella tularensis.
26 c2ywtA_
not modelled
59.9
9
PDB header: transferaseChain: A: PDB Molecule: hypoxanthine-guanine phosphoribosyltransferase;PDBTitle: crystal structure of hypoxanthine-guanine2 phosphoribosyltransferase with gmp from thermus3 thermophilus hb8
27 c3mjdA_
not modelled
59.4
7
PDB header: transferaseChain: A: PDB Molecule: orotate phosphoribosyltransferase;PDBTitle: 1.9 angstrom crystal structure of orotate phosphoribosyltransferase2 (pyre) francisella tularensis.
28 c5vn4A_
not modelled
59.3
12
PDB header: transferaseChain: A: PDB Molecule: adenine phosphoribosyltransferase, putative;PDBTitle: crystal structure of adenine phosphoribosyl transferase from2 trypanosoma brucei in complex with amp, pyrophosphate, and ribose-5-3 phosphate
29 c5eswB_
not modelled
58.9
19
PDB header: transferaseChain: B: PDB Molecule: purine/pyrimidine phosphoribosyltransferase;PDBTitle: crystal structure of apo hypoxanthine-guanine2 phosphoribosyltransferase from legionella pneumophila
30 c2p1zA_
not modelled
56.8
20
PDB header: transferaseChain: A: PDB Molecule: phosphoribosyltransferase;PDBTitle: crystal structure of phosphoribosyltransferase from corynebacterium2 diphtheriae
31 c6apsA_
not modelled
56.2
14
PDB header: transferaseChain: A: PDB Molecule: hypoxanthine-guanine phosphoribosyltransferase;PDBTitle: trypanosoma brucei hypoxanthine guanine phosphoribosyltransferase in2 complex with [(2-((guanine-9h-yl)methyl)propane-1,3 diyl)bis(oxy)3 ]bis(methylene))diphosphonic acid
32 c6fkip_
not modelled
55.6
13
PDB header: membrane proteinChain: P: PDB Molecule: atp synthase subunit c, chloroplastic;PDBTitle: chloroplast f1fo conformation 3
33 c1ychD_
not modelled
54.4
10
PDB header: oxidoreductaseChain: D: PDB Molecule: nitric oxide reductase;PDBTitle: x-ray crystal structures of moorella thermoacetica fpra. novel diiron2 site structure and mechanistic insights into a scavenging nitric3 oxide reductase
34 c3dezA_
not modelled
54.1
15
PDB header: transferaseChain: A: PDB Molecule: orotate phosphoribosyltransferase;PDBTitle: crystal structure of orotate phosphoribosyltransferase from2 streptococcus mutans
35 c3o7mD_
not modelled
53.0
9
PDB header: transferaseChain: D: PDB Molecule: hypoxanthine phosphoribosyltransferase;PDBTitle: 1.98 angstrom resolution crystal structure of a hypoxanthine-guanine2 phosphoribosyltransferase (hpt-2) from bacillus anthracis str. 'ames3 ancestor'
36 c2yzkC_
not modelled
52.9
17
PDB header: transferaseChain: C: PDB Molecule: orotate phosphoribosyltransferase;PDBTitle: crystal structure of orotate phosphoribosyltransferase from2 aeropyrum pernix
37 c4pawA_
not modelled
52.9
10
PDB header: transferaseChain: A: PDB Molecule: orotate phosphoribosyltransferase;PDBTitle: structure of hypothetical protein hp1257.
38 d1vdma1
not modelled
52.3
22
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
39 d1p17b_
not modelled
49.7
17
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
40 d1yfza1
not modelled
49.2
12
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
41 c1yfzA_
not modelled
49.2
12
PDB header: transferaseChain: A: PDB Molecule: hypoxanthine-guanine phosphoribosyltransferase;PDBTitle: novel imp binding in feedback inhibition of hypoxanthine-guanine2 phosphoribosyltransferase from thermoanaerobacter tengcongensis
42 c2przB_
not modelled
46.3
19
PDB header: transferaseChain: B: PDB Molecule: orotate phosphoribosyltransferase 1;PDBTitle: s. cerevisiae orotate phosphoribosyltransferase complexed with omp
43 d1ecfa1
not modelled
44.5
11
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
44 c1vmeB_
not modelled
44.4
10
PDB header: electron transportChain: B: PDB Molecule: flavoprotein;PDBTitle: crystal structure of flavoprotein (tm0755) from thermotoga maritima at2 1.80 a resolution
45 c3m3hA_
not modelled
44.0
20
PDB header: transferaseChain: A: PDB Molecule: orotate phosphoribosyltransferase;PDBTitle: 1.75 angstrom resolution crystal structure of an orotate2 phosphoribosyltransferase from bacillus anthracis str. 'ames3 ancestor'
46 d1qo0d_
not modelled
43.1
14
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: Positive regulator of the amidase operon AmiR
47 d2aeea1
not modelled
41.7
15
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
48 d1hgxa_
not modelled
41.4
9
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
49 d1z7ga1
not modelled
41.3
21
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
50 d1yoba1
not modelled
41.0
17
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related
51 d1tc1a_
not modelled
40.5
15
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
52 c3kb8A_
not modelled
40.2
9
PDB header: transferaseChain: A: PDB Molecule: hypoxanthine phosphoribosyltransferase;PDBTitle: 2.09 angstrom resolution structure of a hypoxanthine-guanine2 phosphoribosyltransferase (hpt-1) from bacillus anthracis str. 'ames3 ancestor' in complex with gmp
53 c5vogA_
not modelled
40.1
17
PDB header: transferaseChain: A: PDB Molecule: putative phosphoribosyltransferase;PDBTitle: crystal structure of a hypothetical protein from neisseria gonorrhoeae2 with bound ppgpp
54 c1pzmB_
not modelled
39.9
13
PDB header: transferaseChain: B: PDB Molecule: hypoxanthine-guanine phosphoribosyltransferase;PDBTitle: crystal structure of hgprt-ase from leishmania tarentolae in complex2 with gmp
55 d1l1qa_
not modelled
38.9
14
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
56 c2iufA_
not modelled
38.7
20
PDB header: oxidoreductaseChain: A: PDB Molecule: catalase;PDBTitle: the structures of penicillium vitale catalase: resting2 state, oxidised state (compound i) and complex with3 aminotriazole
57 c3hlyA_
not modelled
38.1
11
PDB header: flavoproteinChain: A: PDB Molecule: flavodoxin-like domain;PDBTitle: crystal structure of the flavodoxin-like domain from synechococcus sp2 q5mzp6_synp6 protein. northeast structural genomics consortium target3 snr135d.
58 c4zu3D_
not modelled
37.7
24
PDB header: lyaseChain: D: PDB Molecule: halohydrin epoxidase b;PDBTitle: halohydrin hydrogen-halide-lyases, hheb
59 c5ipfA_
not modelled
37.1
12
PDB header: transferaseChain: A: PDB Molecule: hypoxanthine-guanine phosphoribosyltransferase (hgprt);PDBTitle: crystal structure of hypoxanthine-guanine phosphoribosyltransferase2 from schistosoma mansoni in complex with imp
60 c1gph1_
not modelled
37.0
19
PDB header: transferaseChain: 1: PDB Molecule: glutamine phosphoribosyl-pyrophosphate amidotransferase;PDBTitle: structure of the allosteric regulatory enzyme of purine biosynthesis
61 c4rhyC_
not modelled
35.9
11
PDB header: transferase/transferase inhibitorChain: C: PDB Molecule: hypoxanthine-guanine phosphoribosyltransferase;PDBTitle: crystal structures of mycobacterium tuberculosis 6-oxopurine2 phosphoribosyltransferase which is a potential target for drug3 development against this disease
62 d1sqsa_
not modelled
35.3
23
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Hypothetical protein SP1951
63 d1g9sa_
not modelled
35.3
17
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
64 c3mudA_
not modelled
34.9
11
PDB header: contractile proteinChain: A: PDB Molecule: dna repair protein xrcc4,tropomyosin alpha-1 chain;PDBTitle: structure of the tropomyosin overlap complex from chicken smooth2 muscle
65 c3cneD_
not modelled
34.5
9
PDB header: hydrolaseChain: D: PDB Molecule: putative protease i;PDBTitle: crystal structure of the putative protease i from bacteroides2 thetaiotaomicron
66 d1gph11
not modelled
34.0
18
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
67 d1j7ja_
not modelled
33.4
21
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
68 c4rqbB_
not modelled
33.3
12
PDB header: transferaseChain: B: PDB Molecule: hypoxanthine phosphoribosyltransferase;PDBTitle: crystal structure of a hypoxanthine phosphoribosyltransferase (target2 id nysgrc-029686) from staphylococcus aureus (tetragonal space group)
69 c3n2lA_
not modelled
33.1
5
PDB header: transferaseChain: A: PDB Molecule: orotate phosphoribosyltransferase;PDBTitle: 2.1 angstrom resolution crystal structure of an orotate2 phosphoribosyltransferase (pyre) from vibrio cholerae o1 biovar eltor3 str. n16961
70 c4e08B_
not modelled
32.4
23
PDB header: motor proteinChain: B: PDB Molecule: dj-1 beta;PDBTitle: crystal structure of drosophila melanogaster dj-1beta
71 d1ufra_
not modelled
32.0
15
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
72 c2gd7B_
not modelled
31.8
21
PDB header: transcriptionChain: B: PDB Molecule: hexim1 protein;PDBTitle: the structure of the cyclin t-binding domain of hexim12 reveals the molecular basis for regulation of3 transcription elongation
73 c1go4F_
not modelled
31.8
22
PDB header: cell cycleChain: F: PDB Molecule: mitotic spindle assembly checkpoint protein mad1;PDBTitle: crystal structure of mad1-mad2 reveals a conserved mad2 binding motif2 in mad1 and cdc20.
74 d1pzma_
not modelled
31.7
13
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
75 d1qb7a_
not modelled
31.3
20
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
76 c6h9mA_
not modelled
31.1
12
PDB header: membrane proteinChain: A: PDB Molecule: coiled-coil domain-containing protein 90b, mitochondrial,PDBTitle: coiled-coil domain-containing protein 90b residues 43-125 from homo2 sapiens fused to a gcn4 adaptor
77 c4lyyA_
not modelled
30.3
14
PDB header: transferaseChain: A: PDB Molecule: hypoxanthine phosphoribosyltransferase;PDBTitle: crystal structure of hypoxanthine phosphoribosyltransferase from2 shewanella pealeana atcc 700345, nysgrc target 029677.
78 c1ecjB_
not modelled
30.0
11
PDB header: transferaseChain: B: PDB Molecule: glutamine phosphoribosylpyrophosphatePDBTitle: escherichia coli glutamine phosphoribosylpyrophosphate2 (prpp) amidotransferase complexed with 2 amp per tetramer
79 c4z1oB_
not modelled
29.4
24
PDB header: transferaseChain: B: PDB Molecule: phosphoribosyltransferase;PDBTitle: hypoxanthine-guanine-xanthine phosphoribosyltransferase (hgxprt) from2 sulfolobus solfataricus in complex with alpha-3 phosphoribosylpyrophosphoric acid (prpp) and magnesium
80 d2axte1
not modelled
27.7
40
Fold: Single transmembrane helixSuperfamily: Cytochrome b559 subunitsFamily: Cytochrome b559 subunits
81 c3jcue_
not modelled
27.2
33
PDB header: membrane proteinChain: E: PDB Molecule: cytochrome b559 subunit alpha;PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
82 c2wc1A_
not modelled
27.1
17
PDB header: electron transportChain: A: PDB Molecule: flavodoxin;PDBTitle: three-dimensional structure of the nitrogen fixation2 flavodoxin (niff) from rhodobacter capsulatus at 2.2 a
83 c3kziE_
not modelled
26.9
40
PDB header: electron transportChain: E: PDB Molecule: cytochrome b559 subunit alpha;PDBTitle: crystal structure of monomeric form of cyanobacterial photosystem ii
84 c2j5dA_
not modelled
26.9
33
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interacting protein 3;PDBTitle: nmr structure of bnip3 transmembrane domain in lipid bicelles
85 d1p5fa_
not modelled
26.8
19
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: DJ-1/PfpI
86 c3d4oA_
not modelled
26.7
19
PDB header: oxidoreductaseChain: A: PDB Molecule: dipicolinate synthase subunit a;PDBTitle: crystal structure of dipicolinate synthase subunit a (np_243269.1)2 from bacillus halodurans at 2.10 a resolution
87 c4ug1A_
not modelled
26.6
19
PDB header: cell cycleChain: A: PDB Molecule: cell cycle protein gpsb;PDBTitle: gpsb n-terminal domain
88 c5t4oJ_
not modelled
25.4
8
PDB header: hydrolaseChain: J: PDB Molecule: atp synthase subunit b;PDBTitle: autoinhibited e. coli atp synthase state 1
89 d2a4ka1
not modelled
25.3
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases
90 c3qw4B_
not modelled
25.1
22
PDB header: transferase, lyaseChain: B: PDB Molecule: ump synthase;PDBTitle: structure of leishmania donovani ump synthase
91 c3fseB_
not modelled
24.9
26
PDB header: hydrolaseChain: B: PDB Molecule: two-domain protein containing dj-1/thij/pfpi-like andPDBTitle: crystal structure of a two-domain protein containing dj-1/thij/pfpi-2 like and ferritin-like domains (ava_4496) from anabaena variabilis3 atcc 29413 at 1.90 a resolution
92 c4pfqF_
not modelled
24.7
10
PDB header: transferaseChain: F: PDB Molecule: hypoxanthine phosphoribosyltransferase;PDBTitle: crystal structure of hypoxanthine phosphoribosyltransferase from2 brachybacterium faecium dsm 4810, nysgrc target 029763.
93 c5cdjA_
not modelled
24.4
18
PDB header: chaperoneChain: A: PDB Molecule: rubisco large subunit-binding protein subunit alpha,PDBTitle: apical domain of chloroplast chaperonin 60a
94 c2e76E_
not modelled
24.0
26
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
95 c2e75E_
not modelled
24.0
26
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex with 2-nonyl-4-2 hydroxyquinoline n-oxide (nqno) from m.laminosus
96 c1vf5R_
not modelled
24.0
26
PDB header: photosynthesisChain: R: PDB Molecule: protein pet l;PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
97 c2e74E_
not modelled
24.0
26
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex from m.laminosus
98 c1vf5E_
not modelled
24.0
26
PDB header: photosynthesisChain: E: PDB Molecule: protein pet l;PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
99 c4h0lE_
not modelled
24.0
26
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: cytochrome b6f complex crystal structure from mastigocladus laminosus2 with n-side inhibitor nqno
100 d2e74e1
not modelled
24.0
26
Fold: Single transmembrane helixSuperfamily: PetL subunit of the cytochrome b6f complexFamily: PetL subunit of the cytochrome b6f complex
101 c4h13E_
not modelled
24.0
26
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex from mastigocladus2 laminosus with tds
102 c6c4jA_
not modelled
23.0
16
PDB header: oxidoreductaseChain: A: PDB Molecule: udp-glucose 6-dehydrogenase;PDBTitle: ligand bound full length hugdh with a104l substitution
103 c2ka1B_
not modelled
22.9
33
PDB header: membrane proteinChain: B: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles
104 c2ka1A_
not modelled
22.9
33
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles
105 c2ka2A_
not modelled
22.9
33
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles with his173-ser172 intermonomer3 hydrogen bond restraints
106 c2ka2B_
not modelled
22.9
33
PDB header: membrane proteinChain: B: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles with his173-ser172 intermonomer3 hydrogen bond restraints
107 d1srva_
not modelled
22.8
24
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain
108 c2ebuA_
not modelled
22.0
24
PDB header: replicationChain: A: PDB Molecule: replication factor c subunit 1;PDBTitle: solution structure of the brct domain from human2 replication factor c large subunit 1
109 d1uklc_
not modelled
21.9
11
Fold: HLH-likeSuperfamily: HLH, helix-loop-helix DNA-binding domainFamily: HLH, helix-loop-helix DNA-binding domain
110 c4pv1E_
not modelled
21.7
24
PDB header: electron transport/inhibitorChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: cytochrome b6f structure from m. laminosus with the quinone analog2 inhibitor stigmatellin
111 c4i7zE_
not modelled
21.7
24
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of cytochrome b6f in dopg, with disordered rieske2 iron-sulfur protein soluble domain
112 d1sjpa2
not modelled
21.5
22
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain
113 d1oi4a1
not modelled
21.4
19
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: DJ-1/PfpI
114 c2yy0D_
not modelled
21.2
20
PDB header: transcriptionChain: D: PDB Molecule: c-myc-binding protein;PDBTitle: crystal structure of ms0802, c-myc-1 binding protein domain2 from homo sapiens
115 c2jbhA_
not modelled
21.0
19
PDB header: transferaseChain: A: PDB Molecule: phosphoribosyltransferase domain-containing protein 1;PDBTitle: human phosphoribosyl transferase domain containing 1
116 c2e43A_
not modelled
20.5
21
PDB header: transcription/dnaChain: A: PDB Molecule: ccaat/enhancer-binding protein beta;PDBTitle: crystal structure of c/ebpbeta bzip homodimer k269a mutant2 bound to a high affinity dna fragment
117 c4d02A_
not modelled
20.3
8
PDB header: electron transportChain: A: PDB Molecule: anaerobic nitric oxide reductase flavorubredoxin;PDBTitle: the crystallographic structure of flavorubredoxin from escherichia2 coli