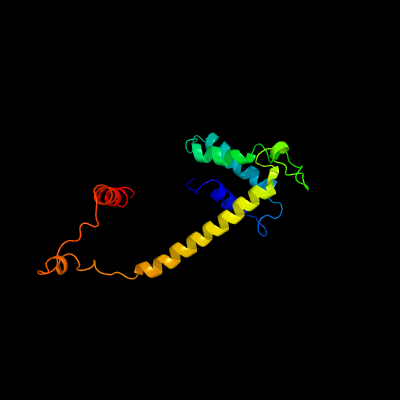
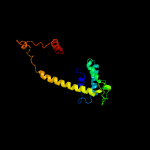
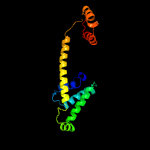
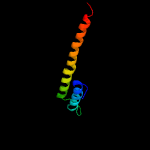
1 c4i98A_
100.0
25
PDB header: cell cycleChain: A: PDB Molecule: segregation and condensation protein a;PDBTitle: crystal structure of the complex between scpa(residues 1-160)-2 scpb(residues 1-183)
2 c3w6jA_
100.0
32
PDB header: cell cycleChain: A: PDB Molecule: scpa;PDBTitle: crystal structure of scpab core complex
3 c3zgxZ_
99.9
40
PDB header: cell cycleChain: Z: PDB Molecule: segregation and condensation protein a;PDBTitle: crystal structure of the kleisin-n smc interface in2 prokaryotic condensin
4 c5h67C_
99.5
25
PDB header: dna binding protein/cell cycleChain: C: PDB Molecule: segregation and condensation protein a;PDBTitle: crystal structure of the bacillus subtilis smc head domain complexed2 with the cognate scpa c-terminal domain and soaked atp
5 c1w1wF_
98.6
18
PDB header: cell adhesionChain: F: PDB Molecule: sister chromatid cohesion protein 1;PDBTitle: sc smc1hd:scc1-c complex, atpgs
6 d1w1we_
98.6
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Rad21/Rec8-like
7 c4i99C_
98.3
32
PDB header: dna binding proteinChain: C: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of the smchead bound to the c-winged helix domain of2 scpa
8 c6qj4E_
86.6
23
PDB header: cell cycleChain: E: PDB Molecule: condensin complex subunit 2;PDBTitle: crystal structure of the c. thermophilum condensin ycs4-brn12 subcomplex bound to the smc4 atpase head in complex with the c-3 terminal domain of brn1
9 c2l01A_
84.1
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of protein bvu3908 from bacteroides vulgatus,2 northeast structural genomics consortium target bvr153
10 c2l02B_
62.8
25
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of protein bt2368 from bacteroides2 thetaiotaomicron, northeast structural genomics consortium target3 btr375
11 c3f41B_
57.5
7
PDB header: hydrolaseChain: B: PDB Molecule: phytase;PDBTitle: structure of the tandemly repeated protein tyrosine2 phosphatase like phytase from mitsuokella multacida
12 d1whqa_
34.5
22
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)
13 c2qdoC_
20.9
21
PDB header: photosynthesisChain: C: PDB Molecule: nbla protein;PDBTitle: nbla protein from t. vulcanus
14 c1u24A_
19.9
15
PDB header: hydrolaseChain: A: PDB Molecule: myo-inositol hexaphosphate phosphohydrolase;PDBTitle: crystal structure of selenomonas ruminantium phytase
15 c2bbrA_
19.9
17
PDB header: viral proteinChain: A: PDB Molecule: viral casp8 and fadd-like apoptosis regulator;PDBTitle: crystal structure of mc159 reveals molecular mechanism of2 disc assembly and vflip inhibition
16 d2gf5a2
18.8
20
Fold: DEATH domainSuperfamily: DEATH domainFamily: DEATH effector domain, DED
17 c2bbzC_
18.1
17
PDB header: viral proteinChain: C: PDB Molecule: viral casp8 and fadd-like apoptosis regulator;PDBTitle: crystal structure of mc159 reveals molecular mechanism of2 disc assembly and vflip inhibition
18 c2ls7A_
16.8
10
PDB header: apoptosisChain: A: PDB Molecule: astrocytic phosphoprotein pea-15;PDBTitle: high definition solution structure of ped/pea-15 death effector domain
19 d1ocka_
14.3
17
Fold: Amidase signature (AS) enzymesSuperfamily: Amidase signature (AS) enzymesFamily: Amidase signature (AS) enzymes
20 d2ixma1
13.8
18
Fold: PTPA-likeSuperfamily: PTPA-likeFamily: PTPA-like
21 c3mhvC_
not modelled
13.7
3
PDB header: protein transportChain: C: PDB Molecule: vacuolar protein sorting-associated protein 4;PDBTitle: crystal structure of vps4 and vta1
22 d1h99a2
not modelled
10.4
18
Fold: PTS-regulatory domain, PRDSuperfamily: PTS-regulatory domain, PRDFamily: PTS-regulatory domain, PRD
23 d1b34b_
not modelled
10.1
27
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP
24 c1b34B_
not modelled
10.1
27
PDB header: rna binding proteinChain: B: PDB Molecule: protein (small nuclear ribonucleoprotein sm d2);PDBTitle: crystal structure of the d1d2 sub-complex from the human snrnp core2 domain
25 c2g62A_
not modelled
9.9
18
PDB header: hydrolase activatorChain: A: PDB Molecule: protein phosphatase 2a, regulatory subunit b' (pr 53);PDBTitle: crystal structure of human ptpa
26 c5h6tB_
not modelled
9.6
13
PDB header: hydrolaseChain: B: PDB Molecule: amidase;PDBTitle: crystal structure of hydrazidase from microbacterium sp. strain hm58-2
27 c4xypA_
not modelled
9.5
15
PDB header: viral proteinChain: A: PDB Molecule: fusion protein;PDBTitle: crystal structure of a piscine viral fusion protein
28 c1ydnA_
not modelled
9.3
17
PDB header: lyaseChain: A: PDB Molecule: hydroxymethylglutaryl-coa lyase;PDBTitle: crystal structure of the hmg-coa lyase from brucella melitensis,2 northeast structural genomics target lr35.
29 d2ozza1
not modelled
9.0
38
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like
30 d1wg8a1
not modelled
8.7
23
Fold: SAM domain-likeSuperfamily: Putative methyltransferase TM0872, insert domainFamily: Putative methyltransferase TM0872, insert domain
31 d1n3ka_
not modelled
8.5
10
Fold: DEATH domainSuperfamily: DEATH domainFamily: DEATH effector domain, DED
32 c3r6uA_
not modelled
8.2
10
PDB header: transport proteinChain: A: PDB Molecule: choline-binding protein;PDBTitle: crystal structure of choline binding protein opubc from bacillus2 subtilis
33 d1m6ya1
not modelled
8.2
30
Fold: SAM domain-likeSuperfamily: Putative methyltransferase TM0872, insert domainFamily: Putative methyltransferase TM0872, insert domain
34 c2l2iB_
not modelled
8.2
86
PDB header: transcriptionChain: B: PDB Molecule: krueppel-like factor 1;PDBTitle: nmr structure of the complex between the tfb1 subunit of tfiih and the2 activation domain of eklf
35 c3pcqX_
not modelled
7.9
44
PDB header: photosynthesisChain: X: PDB Molecule: photosystem i 4.8k protein;PDBTitle: femtosecond x-ray protein nanocrystallography
36 c5j0fA_
not modelled
7.8
32
PDB header: oxidoreductaseChain: A: PDB Molecule: superoxide dismutase [cu-zn],oxidoreductase,superoxidePDBTitle: monomeric human cu,zn superoxide dismutase, loops iv and vii deleted,2 apo form, circular permutant p4/5
37 c5ganm_
not modelled
7.6
20
PDB header: transcriptionChain: M: PDB Molecule: PDBTitle: the overall structure of the yeast spliceosomal u4/u6.u5 tri-snrnp at2 3.7 angstrom
38 c1jb0X_
not modelled
7.2
44
PDB header: photosynthesisChain: X: PDB Molecule: photosystem i subunit psax;PDBTitle: crystal structure of photosystem i: a photosynthetic reaction center2 and core antenna system from cyanobacteria
39 c4fe1X_
not modelled
7.2
44
PDB header: photosynthesisChain: X: PDB Molecule: photosystem i 4.8k protein;PDBTitle: improving the accuracy of macromolecular structure refinement at 7 a2 resolution
40 d1jb0x_
not modelled
7.2
44
Fold: Single transmembrane helixSuperfamily: Subunit PsaX of photosystem I reaction centreFamily: Subunit PsaX of photosystem I reaction centre
41 d2pt0a1
not modelled
7.2
15
Fold: (Phosphotyrosine protein) phosphatases IISuperfamily: (Phosphotyrosine protein) phosphatases IIFamily: Myo-inositol hexaphosphate phosphohydrolase (phytase) PhyA
42 c4z7eB_
not modelled
7.1
10
PDB header: transport proteinChain: B: PDB Molecule: lmo1422 protein;PDBTitle: soluble binding domain of lmo1422 abc-transporter
43 c4tvvA_
not modelled
6.9
10
PDB header: hydrolaseChain: A: PDB Molecule: tyrosine phosphatase ii superfamily protein;PDBTitle: crystal structure of lppa from legionella pneumophila
44 d2azeb1
not modelled
6.8
35
Fold: E2F-DP heterodimerization regionSuperfamily: E2F-DP heterodimerization regionFamily: E2F dimerization segment
45 d1v2za_
not modelled
6.8
15
Fold: KaiA/RbsU domainSuperfamily: KaiA/RbsU domainFamily: Circadian clock protein KaiA, C-terminal domain
46 c1xhoB_
not modelled
6.7
18
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: chorismate mutase;PDBTitle: chorismate mutase from clostridium thermocellum cth-682
47 d1xhoa_
not modelled
6.7
18
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: Chorismate mutase
48 c2gppD_
not modelled
6.5
36
PDB header: transcriptionChain: D: PDB Molecule: nuclear receptor-interacting protein 1;PDBTitle: estrogen related receptor-gamma ligand binding domain2 complexed with a rip140 peptide and synthetic ligand3 gsk4716
49 c4ljoA_
not modelled
6.5
39
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase rnf31;PDBTitle: structure of an active ligase (hoip)/ubiquitin transfer complex
50 c3l6gA_
not modelled
6.3
27
PDB header: glycine betaine-binding proteinChain: A: PDB Molecule: betaine abc transporter permease and substrate bindingPDBTitle: crystal structure of lactococcal opuac in its open conformation
51 c5jqeA_
not modelled
6.3
10
PDB header: hydrolaseChain: A: PDB Molecule: sugar abc transporter substrate-binding protein,caspase-8PDBTitle: crystal structure of caspase8 tded
52 d1fnja_
not modelled
6.3
24
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: Chorismate mutase
53 c5ir0B_
not modelled
6.3
33
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein orf19;PDBTitle: crystal structure of protein of unknown function orf19 from vibrio2 cholerae o1 pici-like element, c57s i109m mutant
54 c2do5A_
not modelled
6.2
38
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: splicing factor 3b subunit 2;PDBTitle: solution structure of the sap domain of human splicing2 factor 3b subunit 2
55 d1sv1a_
not modelled
6.0
15
Fold: KaiA/RbsU domainSuperfamily: KaiA/RbsU domainFamily: Circadian clock protein KaiA, C-terminal domain
56 c5gveA_
not modelled
5.7
16
PDB header: isomerase/protein bindingChain: A: PDB Molecule: dna topoisomerase 3-beta-1;PDBTitle: human top3b-tdrd3 complex
57 d2iboa1
not modelled
5.7
33
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: MTH1187-like
58 c2v6xA_
not modelled
5.7
14
PDB header: protein transportChain: A: PDB Molecule: vacuolar protein sorting-associated protein 4;PDBTitle: stractural insight into the interaction between escrt-iii and vps4
59 c5jwrE_
not modelled
5.7
15
PDB header: transcription regulatorChain: E: PDB Molecule: circadian clock protein kaia;PDBTitle: crystal structure of foldswitch-stabilized kaib in complex with the n-2 terminal ci domain of kaic and a dimer of kaia c-terminal domains3 from thermosynechococcus elongatus
60 c5j2yA_
not modelled
5.6
26
PDB header: gene regulation/dnaChain: A: PDB Molecule: regulatory protein;PDBTitle: molecular insight into the regulatory mechanism of the quorum-sensing2 repressor rsal in pseudomonas aeruginosa
61 d1yqha1
not modelled
5.6
33
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: MTH1187-like
62 c3d7aB_
not modelled
5.5
19
PDB header: unknown functionChain: B: PDB Molecule: upf0201 protein ph1010;PDBTitle: crystal structure of duf54 family protein ph1010 from2 hyperthermophilic archaea pyrococcus horikoshii ot3
63 d1lxna_
not modelled
5.4
27
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: MTH1187-like
64 d1dbfa_
not modelled
5.4
24
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: Chorismate mutase
65 d1pdza1
not modelled
5.2
27
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: Enolase
66 c3bw1A_
not modelled
5.2
33
PDB header: rna binding proteinChain: A: PDB Molecule: u6 snrna-associated sm-like protein lsm3;PDBTitle: crystal structure of homomeric yeast lsm3 exhibiting novel octameric2 ring organisation
67 d1g8fa3
not modelled
5.1
11
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ATP sulfurylase C-terminal domain