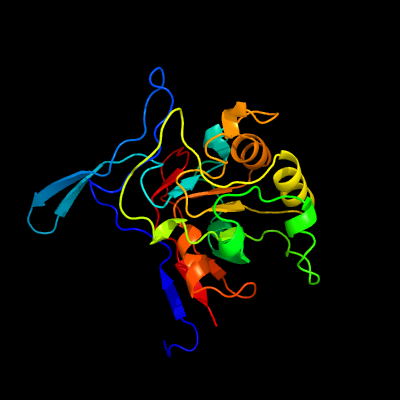
| 1 | c5nnaB_
|
|
 |
100.0 |
26 |
PDB header:hydrolase
Chain: B: PDB Molecule:isatin hydrolase a;
PDBTitle: isatin hydrolase a (iha) from labrenzia aggregata bound to benzyl2 benzoate
|
|
|
|
| 2 | c4j0nA_
|
|
 |
100.0 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:isatin hydrolase b;
PDBTitle: crystal structure of a manganese dependent isatin hydrolase
|
|
|
|
| 3 | c5nmpF_
|
|
 |
100.0 |
25 |
PDB header:hydrolase
Chain: F: PDB Molecule:isatin hydrolase;
PDBTitle: isatin hydrolase a (iha) from ralstonia solanacearum
|
|
|
|
| 4 | c4coaA_
|
|
 |
100.0 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:kynurenine formamidase;
PDBTitle: crystal structure of kynurenine formamidase from bacillus2 anthracis complexed with 2-aminoacetophenone.
|
|
|
|
| 5 | d1r61a_
|
|
 |
100.0 |
27 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Putative cyclase
Family:Putative cyclase |
|
|
|
| 6 | c4cogB_
|
|
 |
100.0 |
26 |
PDB header:hydrolase
Chain: B: PDB Molecule:kynurenine formamidase;
PDBTitle: crystal structure of kynurenine formamidase from burkholderia2 cenocepacia
|
|
|
|
| 7 | c4cobA_
|
|
 |
100.0 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:kynurenine formamidase;
PDBTitle: crystal structure kynurenine formamidase from pseudomonas aeruginosa
|
|
|
|
| 8 | d2b0aa1
|
|
 |
100.0 |
26 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Putative cyclase
Family:Putative cyclase |
|
|
|
| 9 | c5ibzD_
|
|
 |
100.0 |
30 |
PDB header:lyase
Chain: D: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a novel cyclase (pfam04199).
|
|
|
|
| 10 | d2fvta1
|
|
 |
80.6 |
22 |
Fold:MTH938-like
Superfamily:MTH938-like
Family:MTH938-like |
|
|
|
| 11 | c2gm2A_
|
|
 |
57.1 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved hypothetical protein;
PDBTitle: nmr structure of xanthomonas campestris xcc1710: northeast2 structural genomics consortium target xcr35
|
|
|
|
| 12 | d1zy9a2
|
|
 |
46.5 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:YicI catalytic domain-like |
|
|
|
| 13 | c5ir2A_
|
|
 |
42.3 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:cellulase;
PDBTitle: crystal structure of novel cellulases from microbes associated with2 the gut ecosystem
|
|
|
|
| 14 | c2k2eA_
|
|
 |
40.4 |
32 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein bp2786;
PDBTitle: solution nmr structure of bordetella pertussis protein2 bp2786, a mth938-like domain. northeast structural3 genomics consortium target ber31
|
|
|
|
| 15 | c3e4fB_
|
|
 |
39.3 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:aminoglycoside n3-acetyltransferase;
PDBTitle: crystal structure of ba2930- a putative aminoglycoside n3-2 acetyltransferase from bacillus anthracis
|
|
|
|
| 16 | d2nyga1
|
|
 |
38.1 |
13 |
Fold:TTHA0583/YokD-like
Superfamily:TTHA0583/YokD-like
Family:Aminoglycoside 3-N-acetyltransferase-like |
|
|
|
| 17 | c3smaD_
|
|
 |
37.9 |
23 |
PDB header:transferase
Chain: D: PDB Molecule:frbf;
PDBTitle: a new n-acetyltransferase fold in the structure and mechanism of the2 phosphonate biosynthetic enzyme frbf
|
|
|
|
| 18 | c4ce0A_
|
|
 |
37.6 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:o-methyltransferase;
PDBTitle: crystal structure of sah-bound spinosyn rhamnosyl 4'-o-2 methyltransferase spnh from saccharopolyspora spinosa
|
|
|
|
| 19 | d2q4qa1
|
|
 |
37.2 |
13 |
Fold:MTH938-like
Superfamily:MTH938-like
Family:MTH938-like |
|
|
|
| 20 | c4zgwB_
|
|
 |
36.8 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short-chain dehydrogenase/reductase;
PDBTitle: short-chain dehydrogenase/reductase from serratia marcescens bcrc2 10948
|
|
|
|
| 21 | c6bc3A_ |
|
not modelled |
34.5 |
21 |
PDB header:transferase/antibiotic
Chain: A: PDB Molecule:aac 3-vi protein;
PDBTitle: cryo x-ray structure of sisomicin bound aac-via
|
|
|
| 22 | d2fi9a1 |
|
not modelled |
31.6 |
13 |
Fold:MTH938-like
Superfamily:MTH938-like
Family:MTH938-like |
|
|
| 23 | c5ht0B_ |
|
not modelled |
28.9 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:aminoglycoside acetyltransferase hmb0005;
PDBTitle: crystal structure of an antibiotic_nat family aminoglycoside2 acetyltransferase hmb0038 from an uncultured soil metagenomic sample3 in complex with coenzyme a
|
|
|
| 24 | c6mb6A_ |
|
not modelled |
28.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:aac(3)-iiib protein;
PDBTitle: aac-iiib binary with coash
|
|
|
| 25 | c3cvjB_ |
|
not modelled |
28.1 |
8 |
PDB header:isomerase
Chain: B: PDB Molecule:putative phosphoheptose isomerase;
PDBTitle: crystal structure of a putative phosphoheptose isomerase (bh3325) from2 bacillus halodurans c-125 at 2.00 a resolution
|
|
|
| 26 | c1zy9A_ |
|
not modelled |
28.0 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-galactosidase;
PDBTitle: crystal structure of alpha-galactosidase (ec 3.2.1.22) (melibiase)2 (tm1192) from thermotoga maritima at 2.34 a resolution
|
|
|
| 27 | d2gp4a1 |
|
not modelled |
27.5 |
15 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:LeuD/IlvD-like
Family:IlvD/EDD C-terminal domain-like |
|
|
| 28 | c2gp4B_ |
|
not modelled |
24.4 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:6-phosphogluconate dehydratase;
PDBTitle: structure of [fes]cluster-free apo form of 6-phosphogluconate2 dehydratase from shewanella oneidensis
|
|
|
| 29 | c2pcnA_ |
|
not modelled |
23.1 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:s-adenosylmethionine:2-demethylmenaquinone
PDBTitle: crystal structure of s-adenosylmethionine: 2-dimethylmenaquinone2 methyltransferase (gk_1813) from geobacillus kaustophilus hta426
|
|
|
| 30 | c2gp4A_ |
|
not modelled |
22.7 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:6-phosphogluconate dehydratase;
PDBTitle: structure of [fes]cluster-free apo form of 6-phosphogluconate2 dehydratase from shewanella oneidensis
|
|
|
| 31 | c5ykjA_ |
|
not modelled |
22.6 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:peroxiredoxin prx1, mitochondrial;
PDBTitle: structural basis of the thiol resolving mechanism in yeast2 mitochondrial 1-cys peroxiredoxin via glutathione/thioredoxin systems
|
|
|
| 32 | d2bgka1 |
|
not modelled |
21.9 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 33 | c3kw0D_ |
|
not modelled |
20.7 |
17 |
PDB header:hydrolase
Chain: D: PDB Molecule:cysteine peptidase;
PDBTitle: crystal structure of cysteine peptidase (np_982244.1) from bacillus2 cereus atcc 10987 at 2.50 a resolution
|
|
|
| 34 | c3c8oB_ |
|
not modelled |
20.2 |
21 |
PDB header:hydrolase regulator
Chain: B: PDB Molecule:regulator of ribonuclease activity a;
PDBTitle: the crystal structure of rraa from pao1
|
|
|
| 35 | d1mpxa2 |
|
not modelled |
19.9 |
15 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:PepX catalytic domain-like |
|
|
| 36 | c4pf1D_ |
|
not modelled |
18.0 |
15 |
PDB header:hydrolase
Chain: D: PDB Molecule:peptidase s15/coce/nond;
PDBTitle: crystal structure of aminopeptidase from marine sediment archaeon2 thaumarchaeota archaeon
|
|
|
| 37 | c1wu8B_ |
|
not modelled |
18.0 |
15 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein ph0463;
PDBTitle: crystal structure of project ph0463 from pyrococcus horikoshii ot3
|
|
|
| 38 | c2yvaB_ |
|
not modelled |
17.8 |
19 |
PDB header:dna binding protein
Chain: B: PDB Molecule:dnaa initiator-associating protein diaa;
PDBTitle: crystal structure of escherichia coli diaa
|
|
|
| 39 | c2kz3A_ |
|
not modelled |
17.7 |
20 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein rad51l3;
PDBTitle: backbone 1h, 13c, and 15n chemical shift assignments for human rad51d2 from 1 to 83
|
|
|
| 40 | c4xvzB_ |
|
not modelled |
17.2 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:mycinamicin iii 3''-o-methyltransferase;
PDBTitle: mycf mycinamicin iii 3'-o-methyltransferase in complex with mg
|
|
|
| 41 | d1rqpa2 |
|
not modelled |
17.1 |
15 |
Fold:Bacterial fluorinating enzyme, N-terminal domain
Superfamily:Bacterial fluorinating enzyme, N-terminal domain
Family:Bacterial fluorinating enzyme, N-terminal domain |
|
|
| 42 | d2ot2a1 |
|
not modelled |
16.8 |
33 |
Fold:OB-fold
Superfamily:HupF/HypC-like
Family:HupF/HypC-like |
|
|
| 43 | c2eq9C_ |
|
not modelled |
16.6 |
29 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:pyruvate dehydrogenase complex, dihydrolipoamide
PDBTitle: crystal structure of lipoamide dehydrogenase from thermus thermophilus2 hb8 with psbdb
|
|
|
| 44 | c3trjC_ |
|
not modelled |
16.4 |
25 |
PDB header:isomerase
Chain: C: PDB Molecule:phosphoheptose isomerase;
PDBTitle: structure of a phosphoheptose isomerase from francisella tularensis
|
|
|
| 45 | c4gf5S_ |
|
not modelled |
16.4 |
19 |
PDB header:transferase
Chain: S: PDB Molecule:cals11;
PDBTitle: crystal structure of calicheamicin methyltransferase, cals11
|
|
|
| 46 | c2eq8C_ |
|
not modelled |
16.1 |
33 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:pyruvate dehydrogenase complex, dihydrolipoamide
PDBTitle: crystal structure of lipoamide dehydrogenase from thermus thermophilus2 hb8 with psbdp
|
|
|
| 47 | c2q6oB_ |
|
not modelled |
16.0 |
15 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:hypothetical protein;
PDBTitle: sall-y70t with sam and cl
|
|
|
| 48 | d1pjca2 |
|
not modelled |
15.9 |
16 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:L-alanine dehydrogenase-like |
|
|
| 49 | d1q5xa_ |
|
not modelled |
15.2 |
18 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:RraA-like
Family:RraA-like |
|
|
| 50 | c5jc8C_ |
|
not modelled |
15.2 |
24 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative short-chain dehydrogenase/reductase;
PDBTitle: crystal structure of a putative short-chain dehydrogenase/reductase2 from burkholderia xenovorans
|
|
|
| 51 | c2d0iC_ |
|
not modelled |
15.1 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:dehydrogenase;
PDBTitle: crystal structure ph0520 protein from pyrococcus horikoshii ot3
|
|
|
| 52 | c2zbvC_ |
|
not modelled |
14.7 |
23 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:uncharacterized conserved protein;
PDBTitle: crystal structure of uncharacterized conserved protein from thermotoga2 maritima
|
|
|
| 53 | c4xcvA_ |
|
not modelled |
14.7 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadp-dependent 2-hydroxyacid dehydrogenase;
PDBTitle: probable 2-hydroxyacid dehydrogenase from rhizobium etli cfn 42 in2 complex with nadph
|
|
|
| 54 | c1rqrA_ |
|
not modelled |
14.5 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:5'-fluoro-5'-deoxyadenosine synthase;
PDBTitle: crystal structure and mechanism of a bacterial fluorinating enzyme,2 product complex
|
|
|
| 55 | d2d59a1 |
|
not modelled |
14.4 |
28 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
| 56 | c6mn5A_ |
|
not modelled |
14.4 |
14 |
PDB header:transferase/antibiotic
Chain: A: PDB Molecule:aminoglycoside n(3)-acetyltransferase, aac(3)-iva;
PDBTitle: crystal structure of aminoglycoside acetyltransferase aac(3)-iva,2 h154a mutant, in complex with gentamicin c1a
|
|
|
| 57 | c4nx0G_ |
|
not modelled |
14.3 |
25 |
PDB header:hydrolase
Chain: G: PDB Molecule:abp, a gh27 beta-l-arabinopyranosidase;
PDBTitle: crystal structure of abp-wt, a gh27-b-l-arabinopyranosidase from2 geobacillus stearothermophilus
|
|
|
| 58 | d2z1ca1 |
|
not modelled |
14.3 |
60 |
Fold:OB-fold
Superfamily:HupF/HypC-like
Family:HupF/HypC-like |
|
|
| 59 | c4g2nA_ |
|
not modelled |
14.3 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-isomer specific 2-hydroxyacid dehydrogenase, nad-binding;
PDBTitle: crystal structure of putative d-isomer specific 2-hydroxyacid2 dehydrogenase, nad-binding from polaromonas sp. js6 66
|
|
|
| 60 | c3icrA_ |
|
not modelled |
14.1 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:coenzyme a-disulfide reductase;
PDBTitle: crystal structure of oxidized bacillus anthracis coadr-rhd
|
|
|
| 61 | c3d3rA_ |
|
not modelled |
14.1 |
21 |
PDB header:chaperone
Chain: A: PDB Molecule:hydrogenase assembly chaperone hypc/hupf;
PDBTitle: crystal structure of the hydrogenase assembly chaperone hypc/hupf2 family protein from shewanella oneidensis mr-1
|
|
|
| 62 | c1mpxB_ |
|
not modelled |
13.8 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-amino acid ester hydrolase;
PDBTitle: alpha-amino acid ester hydrolase labeled with selenomethionine
|
|
|
| 63 | d2cyja1 |
|
not modelled |
13.8 |
19 |
Fold:MTH938-like
Superfamily:MTH938-like
Family:MTH938-like |
|
|
| 64 | d1tk9a_ |
|
not modelled |
13.7 |
11 |
Fold:SIS domain
Superfamily:SIS domain
Family:mono-SIS domain |
|
|
| 65 | c2f4nA_ |
|
not modelled |
13.7 |
27 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein mj1651;
PDBTitle: crystal structure of protein mj1651 from methanococcus2 jannaschii dsm 2661, pfam duf62
|
|
|
| 66 | c5fohA_ |
|
not modelled |
13.7 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:polysaccharide monooxygenase;
PDBTitle: crystal structure of the catalytic domain of nclpmo9a
|
|
|
| 67 | c5oynB_ |
|
not modelled |
13.6 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:dehydratase, ilvd/edd family;
PDBTitle: crystal structure of d-xylonate dehydratase in holo-form
|
|
|
| 68 | c6ci9D_ |
|
not modelled |
13.5 |
38 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:3-oxoacyl-[acyl-carrier-protein] reductase;
PDBTitle: rmm microcompartment-associated aminopropanol dehydrogenase nadp +2 aminoacetone holo-structure
|
|
|
| 69 | d1j3la_ |
|
not modelled |
13.3 |
17 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:RraA-like
Family:RraA-like |
|
|
| 70 | d2cyua1 |
|
not modelled |
13.1 |
27 |
Fold:Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex
Superfamily:Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex
Family:Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex |
|
|
| 71 | c5u9pB_ |
|
not modelled |
13.1 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:gluconate 5-dehydrogenase;
PDBTitle: crystal structure of a gluconate 5-dehydrogenase from burkholderia2 cenocepacia j2315 in complex with nadp and tartrate
|
|
|
| 72 | c3k4iC_ |
|
not modelled |
12.9 |
20 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein pspto_3204 from2 pseudomonas syringae pv. tomato str. dc3000
|
|
|
| 73 | c2wdzD_ |
|
not modelled |
12.4 |
29 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:short-chain dehydrogenase/reductase;
PDBTitle: crystal structure of the short chain dehydrogenase2 galactitol-dehydrogenase (gatdh) of rhodobacter3 sphaeroides in complex with nad+ and 1,2-pentandiol
|
|
|
| 74 | d1x94a_ |
|
not modelled |
12.3 |
28 |
Fold:SIS domain
Superfamily:SIS domain
Family:mono-SIS domain |
|
|
| 75 | d1vi4a_ |
|
not modelled |
12.0 |
19 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:RraA-like
Family:RraA-like |
|
|
| 76 | c4pcvB_ |
|
not modelled |
11.8 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:bdca (yjgi);
PDBTitle: the structure of bdca (yjgi) from e. coli
|
|
|
| 77 | c4gh5B_ |
|
not modelled |
11.7 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short-chain dehydrogenase/reductase sdr;
PDBTitle: crystal structure of s-2-hydroxypropyl coenzyme m dehydrogenase (s-2 hpcdh)
|
|
|
| 78 | c2cw5B_ |
|
not modelled |
11.6 |
27 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:bacterial fluorinating enzyme homolog;
PDBTitle: crystal structure of a conserved hypothetical protein from2 thermus thermophilus hb8
|
|
|
| 79 | d3d3ra1 |
|
not modelled |
11.5 |
21 |
Fold:OB-fold
Superfamily:HupF/HypC-like
Family:HupF/HypC-like |
|
|
| 80 | c2k4mA_ |
|
not modelled |
11.4 |
25 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0146 protein mth_1000;
PDBTitle: solution nmr structure of m. thermoautotrophicum protein mth_1000,2 northeast structural genomics consortium target tr8
|
|
|
| 81 | c4nbrA_ |
|
not modelled |
11.4 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical 3-oxoacyl-(acyl-carrier protein) reductase;
PDBTitle: crystal structure of 3-oxoacyl-[acyl-carrier protein] reductase from2 brucella melitensis atcc 23457
|
|
|
| 82 | c6nrpC_ |
|
not modelled |
11.2 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:3-oxoacyl-acp reductase fabg;
PDBTitle: putative short-chain dehydrogenase/reductase (sdr) from acinetobacter2 baumannii
|
|
|
| 83 | c3r1iB_ |
|
not modelled |
11.2 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short-chain type dehydrogenase/reductase;
PDBTitle: crystal structure of a short-chain type dehydrogenase/reductase from2 mycobacterium marinum
|
|
|
| 84 | c3es4B_ |
|
not modelled |
11.1 |
33 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein duf861 with a rmlc-like cupin fold;
PDBTitle: crystal structure of protein of unknown function (duf861) with a rmlc-2 like cupin fold (17741406) from agrobacterium tumefaciens str. c583 (dupont) at 1.64 a resolution
|
|
|
| 85 | d2jfga1 |
|
not modelled |
10.8 |
30 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
|
|
| 86 | c5tiiC_ |
|
not modelled |
10.8 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:3-oxoacyl-acp reductase;
PDBTitle: comprehensive analysis of a novel ketoreductase for pentangular2 polyphenol biosynthesis
|
|
|
| 87 | c3rihB_ |
|
not modelled |
10.8 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short chain dehydrogenase or reductase;
PDBTitle: crystal structure of a putative short chain dehydrogenase or reductase2 from mycobacterium abscessus
|
|
|
| 88 | c4fgsB_ |
|
not modelled |
10.6 |
29 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable dehydrogenase protein;
PDBTitle: crystal structure of a probable dehydrogenase protein
|
|
|
| 89 | d2ocda1 |
|
not modelled |
10.4 |
20 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
|
|
| 90 | c4z9yA_ |
|
not modelled |
10.4 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-deoxy-d-gluconate 3-dehydrogenase;
PDBTitle: crystal structure of 2-keto-3-deoxy-d-gluconate dehydrogenase from2 pectobacterium carotovorum
|
|
|
| 91 | c4b79B_ |
|
not modelled |
10.4 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable short-chain dehydrogenase;
PDBTitle: the aeropath project and pseudomonas aeruginosa high-throughput2 crystallographic studies for assessment of potential targets in3 early stage drug discovery.
|
|
|
| 92 | c2wk1A_ |
|
not modelled |
10.3 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:novp;
PDBTitle: structure of the o-methyltransferase novp
|
|
|
| 93 | c3awdD_ |
|
not modelled |
10.3 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:putative polyol dehydrogenase;
PDBTitle: crystal structure of gox2181
|
|
|
| 94 | d2c42a4 |
|
not modelled |
10.3 |
21 |
Fold:Pyruvate-ferredoxin oxidoreductase, PFOR, domain III
Superfamily:Pyruvate-ferredoxin oxidoreductase, PFOR, domain III
Family:Pyruvate-ferredoxin oxidoreductase, PFOR, domain III |
|
|
| 95 | c1nxjA_ |
|
not modelled |
10.2 |
18 |
PDB header:unknown function
Chain: A: PDB Molecule:probable s-adenosylmethionine:2-
PDBTitle: structure of rv3853 from mycobacterium tuberculosis
|
|
|
| 96 | d1nxja_ |
|
not modelled |
10.2 |
18 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:RraA-like
Family:RraA-like |
|
|
| 97 | d2obba1 |
|
not modelled |
10.2 |
19 |
Fold:HAD-like
Superfamily:HAD-like
Family:BT0820-like |
|
|
| 98 | d1ygya1 |
|
not modelled |
10.2 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
|
|
| 99 | d1w85i_ |
|
not modelled |
10.0 |
42 |
Fold:Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex
Superfamily:Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex
Family:Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex |
|
|