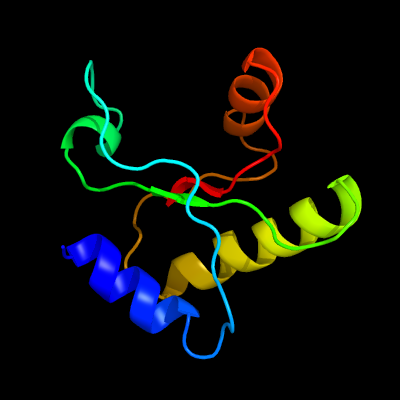
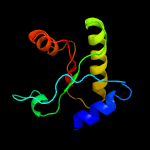
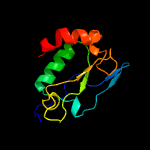
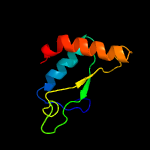
1 d1lbua2
95.9
29
Fold: Hedgehog/DD-peptidaseSuperfamily: Hedgehog/DD-peptidaseFamily: Muramoyl-pentapeptide carboxypeptidase
2 c4ox3A_
95.0
15
PDB header: hydrolaseChain: A: PDB Molecule: putative carboxypeptidase yodj;PDBTitle: structure of the ldcb ld-carboxypeptidase reveals the molecular basis2 of peptidoglycan recognition
3 c5hnmC_
94.7
27
PDB header: hydrolaseChain: C: PDB Molecule: d-alanyl-d-alanine carboxypeptidase;PDBTitle: crystal structure of vancomycin resistance d,d-pentapeptidase vany2 e175a mutant from vanb-type resistance cassette in complex with3 zn(ii)
4 c4murA_
94.4
26
PDB header: hydrolaseChain: A: PDB Molecule: d,d-dipeptidase/d,d-carboxypeptidase;PDBTitle: crystal structure of vancomycin resistance d,d-dipeptidase/d,d-2 pentapeptidase vanxyc d59s mutant
5 c4ox5A_
94.2
27
PDB header: hydrolaseChain: A: PDB Molecule: ldcb ld-carboxypeptidase;PDBTitle: structure of the ldcb ld-carboxypeptidase reveals the molecular basis2 of peptidoglycan recognition
6 c1lbuA_
94.1
22
PDB header: hydrolaseChain: A: PDB Molecule: muramoyl-pentapeptide carboxypeptidase;PDBTitle: hydrolase metallo (zn) dd-peptidase
7 d2vo9a1
93.2
23
Fold: Hedgehog/DD-peptidaseSuperfamily: Hedgehog/DD-peptidaseFamily: VanY-like
8 c2vo9C_
91.2
22
PDB header: hydrolaseChain: C: PDB Molecule: l-alanyl-d-glutamate peptidase;PDBTitle: crystal structure of the enzymatically active domain of the listeria2 monocytogenes bacteriophage 500 endolysin ply500
9 c4jidA_
88.6
24
PDB header: hydrolaseChain: A: PDB Molecule: d-alanyl-d-alanine carboxypeptidase family protein;PDBTitle: crystal structure of baldcb / vany-like l,d-carboxypeptidase zinc(ii)-2 free
10 c4f78A_
87.7
22
PDB header: hydrolaseChain: A: PDB Molecule: d,d-dipeptidase/d,d-carboxypeptidase;PDBTitle: crystal structure of vancomycin resistance d,d-dipeptidase vanxyg
11 d1tzpa_
87.3
17
Fold: Hedgehog/DD-peptidaseSuperfamily: Hedgehog/DD-peptidaseFamily: MepA-like
12 c5opzB_
86.4
24
PDB header: hydrolaseChain: B: PDB Molecule: chix;PDBTitle: crystal structure of serratia marcescens l-ala d-glu endopeptidase2 chix
13 c2mxzA_
82.6
24
PDB header: hydrolaseChain: A: PDB Molecule: l-alanyl-d-glutamate peptidase;PDBTitle: bacteriophage t5 l-alanoyl-d-glutamate peptidase comlpex with zn2+2 (endo t5-zn2+)
14 d3d1ma1
78.3
22
Fold: Hedgehog/DD-peptidaseSuperfamily: Hedgehog/DD-peptidaseFamily: Hedgehog (development protein), N-terminal signaling domain
15 c3m1nB_
61.5
22
PDB header: signaling proteinChain: B: PDB Molecule: sonic hedgehog protein;PDBTitle: crystal structure of human sonic hedgehog n-terminal domain
16 d2ibge1
46.6
19
Fold: Hedgehog/DD-peptidaseSuperfamily: Hedgehog/DD-peptidaseFamily: Hedgehog (development protein), N-terminal signaling domain
17 d1pbaa_
38.6
7
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Pancreatic carboxypeptidase, activation domain
18 c3rcnA_
29.7
15
PDB header: hydrolaseChain: A: PDB Molecule: beta-n-acetylhexosaminidase;PDBTitle: crystal structure of beta-n-acetylhexosaminidase from arthrobacter2 aurescens
19 c5h4uB_
26.6
45
PDB header: hydrolaseChain: B: PDB Molecule: endo-beta-1,4-glucanase;PDBTitle: crystal structure of cellulase from antarctic springtail, cryptopygus2 antarcticus
20 d1kwma2
24.4
10
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Pancreatic carboxypeptidase, activation domain
21 d1nsaa2
not modelled
22.8
7
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Pancreatic carboxypeptidase, activation domain
22 c2zhhA_
not modelled
19.2
23
PDB header: transcriptionChain: A: PDB Molecule: redox-sensitive transcriptional activator soxr;PDBTitle: crystal structure of soxr
23 c6q63B_
not modelled
15.7
15
PDB header: hydrolaseChain: B: PDB Molecule: beta-hexosaminidase;PDBTitle: bt0459
24 d1o6xa_
not modelled
15.6
20
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Pancreatic carboxypeptidase, activation domain
25 d1ayea2
not modelled
15.2
20
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Pancreatic carboxypeptidase, activation domain
26 c4i8oA_
not modelled
13.5
12
PDB header: toxinChain: A: PDB Molecule: toxin rnla;PDBTitle: crystal structure of the toxin rnla from escherichia coli
27 d1pyta_
not modelled
13.5
23
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Pancreatic carboxypeptidase, activation domain
28 d1yloa2
not modelled
13.4
19
Fold: Phosphorylase/hydrolase-likeSuperfamily: Zn-dependent exopeptidasesFamily: Bacterial dinuclear zinc exopeptidases
29 c2fvgA_
not modelled
13.0
12
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase;PDBTitle: crystal structure of endoglucanase (tm1049) from thermotoga maritima2 at 2.01 a resolution
30 c2k5kA_
not modelled
12.8
36
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein rhr2;PDBTitle: solution structure of rhr2 from rhodobacter sphaeroides.2 northeast structural genomics consortium
31 d2fhfa2
not modelled
12.6
26
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes
32 d1pcaa1
not modelled
12.2
27
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Pancreatic carboxypeptidase, activation domain
33 d1jqga2
not modelled
11.7
20
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Pancreatic carboxypeptidase, activation domain
34 c5j4aA_
not modelled
11.6
16
PDB header: toxinChain: A: PDB Molecule: trna nuclease cdia;PDBTitle: cdia-ct toxin from burkholderia pseudomallei e479 in complex with2 cognate cdii immunity protein
35 d1l8fa_
not modelled
10.5
40
Fold: Double psi beta-barrelSuperfamily: Barwin-like endoglucanasesFamily: Eng V-like
36 d2boaa2
not modelled
9.3
16
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Pancreatic carboxypeptidase, activation domain
37 c3rpcD_
not modelled
9.1
40
PDB header: hydrolaseChain: D: PDB Molecule: possible metal-dependent hydrolase;PDBTitle: the crystal structure of a possible metal-dependent hydrolase from2 veillonella parvula dsm 2008
38 c1qbaA_
not modelled
8.7
26
PDB header: glycosyl hydrolaseChain: A: PDB Molecule: chitobiase;PDBTitle: bacterial chitobiase, glycosyl hydrolase family 20
39 c9rubB_
not modelled
8.5
26
PDB header: lyase(carbon-carbon)Chain: B: PDB Molecule: ribulose-1,5-bisphosphate carboxylase;PDBTitle: crystal structure of activated ribulose-1,5-bisphosphate carboxylase2 complexed with its substrate, ribulose-1,5-bisphosphate
40 c6ezrA_
not modelled
8.5
21
PDB header: hydrolaseChain: A: PDB Molecule: beta-n-acetylglucosaminidase nag2;PDBTitle: crystal structure of gh20 exo beta-n-acetylglucosaminidase from vibrio2 harveyi
41 d1wiga2
not modelled
8.3
38
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain
42 d2cg7a1
not modelled
8.3
63
Fold: FnI-like domainSuperfamily: FnI-like domainFamily: Fibronectin type I module
43 c5c5zA_
not modelled
8.0
30
PDB header: hydrolaseChain: A: PDB Molecule: glutamyl-trna amidotransferase;PDBTitle: crystal structure analysis of c4763, a uropathogenic e. coli-specific2 protein
44 c4jo0A_
not modelled
7.9
50
PDB header: oxidoreductaseChain: A: PDB Molecule: cmla;PDBTitle: crystal structure of cmla, a diiron beta-hydroxylase from streptomyces2 venezuelae
45 c4qn9A_
not modelled
7.7
50
PDB header: hydrolaseChain: A: PDB Molecule: n-acyl-phosphatidylethanolamine-hydrolyzing phospholipasePDBTitle: structure of human nape-pld
46 c4hlmD_
not modelled
7.6
45
PDB header: transferaseChain: D: PDB Molecule: tankyrase-2;PDBTitle: crystal structure of tankyrase 2 in complex with 3',4'-2 dihydroxyflavone
47 c6b4hB_
not modelled
7.6
40
PDB header: transport proteinChain: B: PDB Molecule: nucleoporin amo1;PDBTitle: crystal structure of chaetomium thermophilum gle1 ctd-nup42 gbm-ip62 complex
48 c2rkzD_
not modelled
7.5
63
PDB header: cell adhesionChain: D: PDB Molecule: fibronectin;PDBTitle: crystal structure of the second and third fibronectin f1 modules in2 complex with a fragment of staphylococcus aureus fnbpa-1
49 d1r44a_
not modelled
7.4
24
Fold: Hedgehog/DD-peptidaseSuperfamily: Hedgehog/DD-peptidaseFamily: VanX-like
50 c3k44D_
not modelled
7.3
50
PDB header: nucleic acid binding proteinChain: D: PDB Molecule: purine-rich binding protein-alpha, isoform b;PDBTitle: crystal structure of drosophila melanogaster pur-alpha
51 d1nowa1
not modelled
6.9
16
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-N-acetylhexosaminidase catalytic domain
52 c2fekA_
not modelled
6.6
33
PDB header: hydrolaseChain: A: PDB Molecule: low molecular weight protein-tyrosine-PDBTitle: structure of a protein tyrosine phosphatase
53 c5fgoA_
not modelled
6.3
40
PDB header: dna binding proteinChain: A: PDB Molecule: cg1507-pb, isoform b;PDBTitle: crystal structure of d. melanogaster pur-alpha repeat iii.
54 d1ug0a_
not modelled
6.2
6
Fold: Surp module (SWAP domain)Superfamily: Surp module (SWAP domain)Family: Surp module (SWAP domain)
55 c3qb8A_
not modelled
6.1
12
PDB header: transferaseChain: A: PDB Molecule: a654l protein;PDBTitle: paramecium chlorella bursaria virus1 putative orf a654l is a polyamine2 acetyltransferase
56 d2rkya2
not modelled
6.1
63
Fold: FnI-like domainSuperfamily: FnI-like domainFamily: Fibronectin type I module
57 c2wylF_
not modelled
6.0
44
PDB header: hydrolaseChain: F: PDB Molecule: l-ascorbate-6-phosphate lactonase ulag;PDBTitle: apo structure of a metallo-b-lactamase
58 c2imuA_
not modelled
5.8
44
PDB header: viral proteinChain: A: PDB Molecule: structural polyprotein (pp) p1;PDBTitle: nmr structure of pep46 from the infectious bursal disease2 virus (ibdv) in dodecylphosphocholin (dpc).
59 d1y1la_
not modelled
5.8
63
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: Phosphotyrosine protein phosphatases IFamily: Low-molecular-weight phosphotyrosine protein phosphatases
60 d2rkya1
not modelled
5.7
50
Fold: FnI-like domainSuperfamily: FnI-like domainFamily: Fibronectin type I module
61 c2rkyA_
not modelled
5.7
63
PDB header: cell adhesionChain: A: PDB Molecule: fibronectin;PDBTitle: crystal structure of the fourth and fifth fibronectin f1 modules in2 complex with a fragment of staphylococcus aureus fnbpa-1
62 c2k89A_
not modelled
5.7
40
PDB header: protein bindingChain: A: PDB Molecule: phospholipase a-2-activating protein;PDBTitle: solution structure of a novel ubiquitin-binding domain from2 human plaa (pfuc, gly76-pro77 cis isomer)
63 c5i8iD_
not modelled
5.5
27
PDB header: hydrolaseChain: D: PDB Molecule: urea amidolyase;PDBTitle: crystal structure of the k. lactis urea amidolyase
64 c4lrqC_
not modelled
5.5
56
PDB header: hydrolaseChain: C: PDB Molecule: phosphotyrosine protein phosphatase;PDBTitle: crystal structure of a low molecular weight phosphotyrosine2 phosphatase from vibrio choleraeo395
65 c3w36A_
not modelled
5.4
30
PDB header: oxidoreductaseChain: A: PDB Molecule: naph1;PDBTitle: crystal structure of holo-type bacterial vanadium-dependent2 chloroperoxidase
66 c3pb6X_
not modelled
5.4
12
PDB header: transferaseChain: X: PDB Molecule: glutaminyl-peptide cyclotransferase-like protein;PDBTitle: crystal structure of the catalytic domain of human golgi-resident2 glutaminyl cyclase at ph 6.5
67 c3bv6D_
not modelled
5.3
33
PDB header: hydrolaseChain: D: PDB Molecule: metal-dependent hydrolase;PDBTitle: crystal structure of uncharacterized metallo protein from vibrio2 cholerae with beta-lactamase like fold
68 d1t1ua1
not modelled
5.1
26
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: Choline/Carnitine O-acyltransferase