| 1 |
|
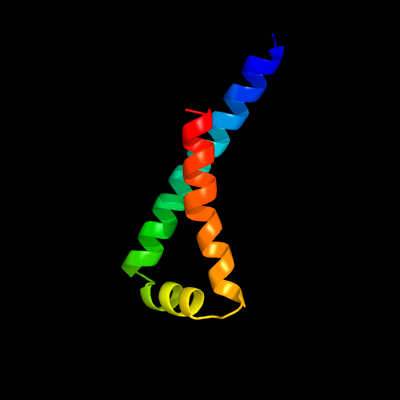
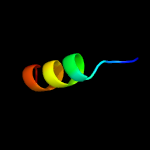
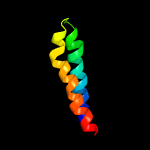
PDB 3m7b chain A
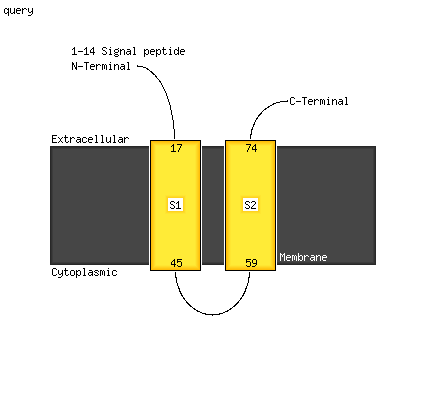
Region: 15 - 77
Aligned: 63
Modelled: 63
Confidence: 98.4%
Identity: 16%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:tellurite resistance protein teha homolog;
PDBTitle: crystal structure of plant slac1 homolog teha
Phyre2
| 2 |
|
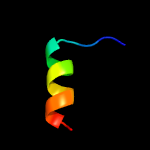
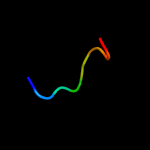
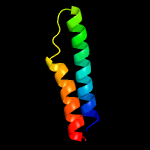
PDB 3m7e chain A
Region: 15 - 77
Aligned: 63
Modelled: 63
Confidence: 98.4%
Identity: 16%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:tellurite resistance protein teha homolog;
PDBTitle: crystal structure of plant slac1 homolog teha
Phyre2
| 3 |
|
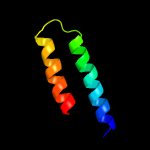
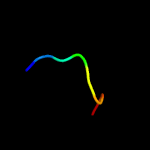
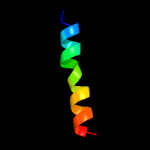
PDB 5cof chain A
Region: 2 - 16
Aligned: 15
Modelled: 15
Confidence: 12.9%
Identity: 27%
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterised protein q1r1x2 from escherichia2 coli uti89
Phyre2
| 4 |
|
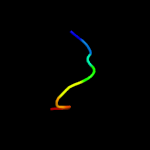
PDB 6cfw chain B
Region: 25 - 79
Aligned: 41
Modelled: 41
Confidence: 10.5%
Identity: 24%
PDB header:membrane protein
Chain: B: PDB Molecule:monovalent cation/h+ antiporter subunit f;
PDBTitle: cryoem structure of a respiratory membrane-bound hydrogenase
Phyre2
| 5 |
|
PDB 2a6a chain A domain 1
Region: 59 - 65
Aligned: 7
Modelled: 7
Confidence: 9.8%
Identity: 43%
Fold: Ribonuclease H-like motif
Superfamily: Actin-like ATPase domain
Family: YeaZ-like
Phyre2
| 6 |
|
PDB 6n9a chain B
Region: 58 - 65
Aligned: 8
Modelled: 8
Confidence: 9.4%
Identity: 38%
PDB header:biosynthetic protein
Chain: B: PDB Molecule:trna threonylcarbamoyladenosine biosynthesis protein tsab;
PDBTitle: crystal structure of thermotoga maritima threonylcarbamoyladenosine2 biosynthesis complex tsab2d2e2 bound to atp and carboxy-amp
Phyre2
| 7 |
|
PDB 6anr chain A
Region: 2 - 16
Aligned: 15
Modelled: 15
Confidence: 8.8%
Identity: 27%
PDB header:hydrolase
Chain: A: PDB Molecule:colibactin self-protection protein clbs;
PDBTitle: crystal structure of a self resistance protein clbs from colibactin2 biosynthetic gene cluster
Phyre2
| 8 |
|
PDB 2lor chain A
Region: 62 - 72
Aligned: 11
Modelled: 11
Confidence: 8.6%
Identity: 45%
PDB header:membrane protein
Chain: A: PDB Molecule:transmembrane protein 141;
PDBTitle: backbone structure of human membrane protein tmem141
Phyre2
| 9 |
|
PDB 1okj chain B
Region: 58 - 65
Aligned: 8
Modelled: 8
Confidence: 7.6%
Identity: 63%
PDB header:hydrolase
Chain: B: PDB Molecule:trna threonylcarbamoyladenosine biosynthesis protein tsab;
PDBTitle: crystal structure of the essential e. coli yeaz2 protein by mad method using the gadolinium complex3 "dotma"
Phyre2
| 10 |
|
PDB 1okj chain A domain 1
Region: 59 - 65
Aligned: 7
Modelled: 7
Confidence: 7.4%
Identity: 71%
Fold: Ribonuclease H-like motif
Superfamily: Actin-like ATPase domain
Family: YeaZ-like
Phyre2
| 11 |
|
PDB 5br9 chain C
Region: 58 - 65
Aligned: 8
Modelled: 8
Confidence: 7.3%
Identity: 63%
PDB header:unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of an uncharacterized protein with similarity to2 peptidase yeaz from pseudomonas aeruginosa
Phyre2
| 12 |
|
PDB 3r6m chain D
Region: 58 - 65
Aligned: 8
Modelled: 8
Confidence: 7.3%
Identity: 63%
PDB header:hydrolase
Chain: D: PDB Molecule:yeaz, resuscitation promoting factor;
PDBTitle: crystal structure of vibrio parahaemolyticus yeaz
Phyre2
| 13 |
|
PDB 2a6a chain B
Region: 58 - 65
Aligned: 8
Modelled: 8
Confidence: 6.6%
Identity: 38%
PDB header:hydrolase
Chain: B: PDB Molecule:hypothetical protein tm0874;
PDBTitle: crystal structure of glycoprotein endopeptidase (tm0874) from2 thermotoga maritima at 2.50 a resolution
Phyre2
| 14 |
|
PDB 3dww chain A
Region: 28 - 78
Aligned: 50
Modelled: 51
Confidence: 6.5%
Identity: 10%
PDB header:isomerase
Chain: A: PDB Molecule:prostaglandin e synthase;
PDBTitle: electron crystallographic structure of human microsomal prostaglandin2 e synthase 1
Phyre2
| 15 |
|
PDB 3leo chain A
Region: 6 - 75
Aligned: 47
Modelled: 49
Confidence: 6.4%
Identity: 23%
PDB header:lyase
Chain: A: PDB Molecule:leukotriene c4 synthase;
PDBTitle: structure of human leukotriene c4 synthase mutant r31q in complex with2 glutathione
Phyre2
| 16 |
|
PDB 3oh8 chain A
Region: 39 - 59
Aligned: 21
Modelled: 21
Confidence: 5.9%
Identity: 24%
PDB header:isomerase
Chain: A: PDB Molecule:nucleoside-diphosphate sugar epimerase (sula family);
PDBTitle: crystal structure of the nucleoside-diphosphate sugar epimerase from2 corynebacterium glutamicum. northeast structural genomics consortium3 target cgr91
Phyre2