| 1 |
|
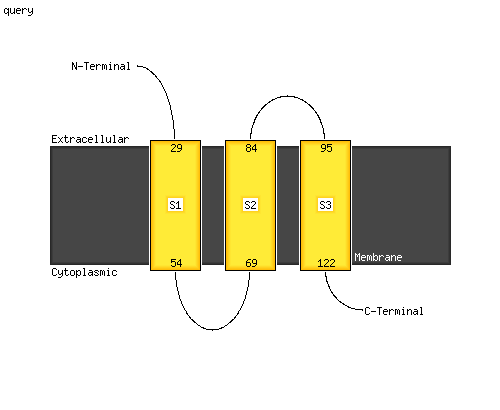
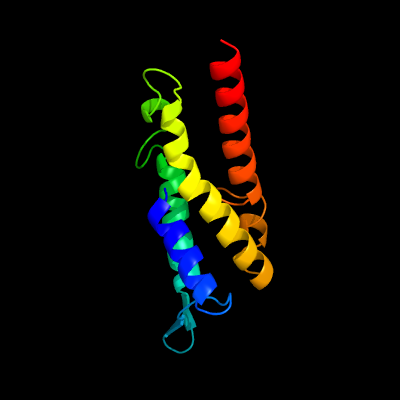
PDB 3m7b chain A
Region: 1 - 123
Aligned: 104
Modelled: 123
Confidence: 99.7%
Identity: 16%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:tellurite resistance protein teha homolog;
PDBTitle: crystal structure of plant slac1 homolog teha
Phyre2
| 2 |
|
PDB 3m7e chain A
Region: 1 - 123
Aligned: 104
Modelled: 120
Confidence: 99.7%
Identity: 16%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:tellurite resistance protein teha homolog;
PDBTitle: crystal structure of plant slac1 homolog teha
Phyre2
| 3 |
|
PDB 5klc chain A
Region: 127 - 155
Aligned: 29
Modelled: 29
Confidence: 49.5%
Identity: 28%
PDB header:sugar binding protein
Chain: A: PDB Molecule:carbohydrate binding module e1;
PDBTitle: structure of cbm_e1, a novel carbohydrate-binding module found by2 sugar cane soil metagenome
Phyre2
| 4 |
|
PDB 6adq chain P
Region: 70 - 127
Aligned: 58
Modelled: 58
Confidence: 20.3%
Identity: 12%
PDB header:electron transport
Chain: P: PDB Molecule:prokaryotic respiratory supercomplex associate factor 1
PDBTitle: respiratory complex ciii2civ2sod2 from mycobacterium smegmatis
Phyre2
| 5 |
|
PDB 6hu9 chain T
Region: 58 - 72
Aligned: 15
Modelled: 15
Confidence: 15.8%
Identity: 13%
PDB header:oxidoreductase/electron transport
Chain: T: PDB Molecule:cytochrome b-c1 complex subunit 9;
PDBTitle: iii2-iv2 mitochondrial respiratory supercomplex from s. cerevisiae
Phyre2
| 6 |
|
PDB 2ciw chain A domain 2
Region: 54 - 69
Aligned: 16
Modelled: 16
Confidence: 15.3%
Identity: 19%
Fold: EF Hand-like
Superfamily: Cloroperoxidase
Family: Cloroperoxidase
Phyre2
| 7 |
|
PDB 2rrf chain A
Region: 108 - 132
Aligned: 25
Modelled: 25
Confidence: 11.8%
Identity: 24%
PDB header:unknown function
Chain: A: PDB Molecule:zinc finger fyve domain-containing protein 21;
PDBTitle: the solution structure of the c-terminal region of zinc finger fyve2 domain-containing protein 21
Phyre2
| 8 |
|
PDB 1z65 chain A
Region: 41 - 51
Aligned: 11
Modelled: 11
Confidence: 10.2%
Identity: 27%
PDB header:unknown function
Chain: A: PDB Molecule:prion-like protein doppel;
PDBTitle: mouse doppel 1-30 peptide
Phyre2
| 9 |
|
PDB 4hxh chain C
Region: 150 - 161
Aligned: 12
Modelled: 12
Confidence: 8.8%
Identity: 33%
PDB header:rna/rna binding protein/hydrolase
Chain: C: PDB Molecule:histone rna hairpin-binding protein;
PDBTitle: structure of mrna stem-loop, human stem-loop binding protein and2 3'hexo ternary complex
Phyre2
| 10 |
|
PDB 2k1a chain A
Region: 159 - 164
Aligned: 6
Modelled: 6
Confidence: 8.5%
Identity: 67%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
Phyre2
| 11 |
|
PDB 5yq7 chain L
Region: 25 - 132
Aligned: 108
Modelled: 108
Confidence: 7.2%
Identity: 13%
PDB header:photosynthesis
Chain: L: PDB Molecule:precursor for l subunits of photosynthetic reaction center;
PDBTitle: cryo-em structure of the rc-lh core complex from roseiflexus2 castenholzii
Phyre2
| 12 |
|
PDB 2bjq chain A domain 2
Region: 124 - 150
Aligned: 27
Modelled: 27
Confidence: 7.0%
Identity: 22%
Fold: MFPT repeat-like
Superfamily: MFPT repeat-like
Family: MFPT repeat
Phyre2
| 13 |
|
PDB 5ldw chain C
Region: 28 - 40
Aligned: 13
Modelled: 13
Confidence: 6.7%
Identity: 46%
PDB header:oxidoreductase
Chain: C: PDB Molecule:nadh dehydrogenase [ubiquinone] iron-sulfur protein 3,
PDBTitle: structure of mammalian respiratory complex i, class1
Phyre2
| 14 |
|
PDB 5lc5 chain C
Region: 28 - 40
Aligned: 13
Modelled: 13
Confidence: 6.7%
Identity: 46%
PDB header:oxidoreductase
Chain: C: PDB Molecule:nadh dehydrogenase [ubiquinone] iron-sulfur protein 3,
PDBTitle: structure of mammalian respiratory complex i, class2
Phyre2
| 15 |
|
PDB 6c24 chain A
Region: 141 - 150
Aligned: 10
Modelled: 10
Confidence: 6.6%
Identity: 60%
PDB header:gene regulation
Chain: A: PDB Molecule:polycomb protein suz12;
PDBTitle: cryo-em structure of prc2 bound to cofactors aebp2 and jarid2 in the2 extended active state
Phyre2
| 16 |
|
PDB 2p5m chain A domain 1
Region: 137 - 161
Aligned: 25
Modelled: 25
Confidence: 6.5%
Identity: 32%
Fold: DCoH-like
Superfamily: C-terminal domain of arginine repressor
Family: C-terminal domain of arginine repressor
Phyre2
| 17 |
|
PDB 5a63 chain D
Region: 96 - 114
Aligned: 19
Modelled: 19
Confidence: 5.6%
Identity: 21%
PDB header:hydrolase
Chain: D: PDB Molecule:gamma-secretase subunit pen-2;
PDBTitle: cryo-em structure of the human gamma-secretase complex at 3.4 angstrom2 resolution.
Phyre2
| 18 |
|
PDB 2v4j chain A domain 2
Region: 146 - 164
Aligned: 18
Modelled: 19
Confidence: 5.4%
Identity: 44%
Fold: Ferredoxin-like
Superfamily: Nitrite/Sulfite reductase N-terminal domain-like
Family: DsrA/DsrB N-terminal-domain-like
Phyre2
| 19 |
|
PDB 3ayg chain A
Region: 25 - 131
Aligned: 102
Modelled: 107
Confidence: 5.3%
Identity: 12%
PDB header:oxidoreductase
Chain: A: PDB Molecule:nitric oxide reductase;
PDBTitle: crystal structure of nitric oxide reductase complex with hqno
Phyre2
| 20 |
|
PDB 1xxa chain A
Region: 137 - 161
Aligned: 25
Modelled: 25
Confidence: 5.1%
Identity: 20%
Fold: DCoH-like
Superfamily: C-terminal domain of arginine repressor
Family: C-terminal domain of arginine repressor
Phyre2