1 c2qa2A_
100.0
21
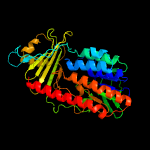
PDB header: oxidoreductaseChain: A: PDB Molecule: polyketide oxygenase cabe;PDBTitle: crystal structure of cabe, an aromatic hydroxylase from angucycline2 biosynthesis, determined to 2.7 a resolution
2 c2qa1A_
100.0
22
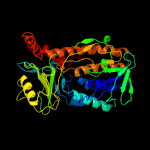
PDB header: oxidoreductaseChain: A: PDB Molecule: polyketide oxygenase pgae;PDBTitle: crystal structure of pgae, an aromatic hydroxylase involved in2 angucycline biosynthesis
3 c6j0zC_
100.0
20
PDB header: oxidoreductaseChain: C: PDB Molecule: putative angucycline-like polyketide oxygenase;PDBTitle: crystal structure of alpk
4 c2dkhA_
100.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: 3-hydroxybenzoate hydroxylase;PDBTitle: crystal structure of 3-hydroxybenzoate hydroxylase from comamonas2 testosteroni, in complex with the substrate
5 c4k2xB_
100.0
22
PDB header: oxidoreductase, flavoproteinChain: B: PDB Molecule: polyketide oxygenase/hydroxylase;PDBTitle: oxys anhydrotetracycline hydroxylase from streptomyces rimosus
6 c3fmwC_
100.0
23
PDB header: oxidoreductaseChain: C: PDB Molecule: oxygenase;PDBTitle: the crystal structure of mtmoiv, a baeyer-villiger monooxygenase from2 the mithramycin biosynthetic pathway in streptomyces argillaceus.
7 c5kowA_
100.0
23
PDB header: oxidoreductaseChain: A: PDB Molecule: pentachlorophenol 4-monooxygenase;PDBTitle: structure of rifampicin monooxygenase
8 c6c6rA_
100.0
14
PDB header: oxidoreductase/oxidoreductase inhibitorChain: A: PDB Molecule: squalene monooxygenase;PDBTitle: human squalene epoxidase (sqle, squalene monooxygenase) structure with2 fad
9 c5xgvB_
100.0
22
PDB header: oxidoreductaseChain: B: PDB Molecule: pyre3;PDBTitle: the structure of diels-alderase pyre3 in the biosynthetic pathway of2 pyrroindomycins
10 c3i3lA_
100.0
20
PDB header: hydrolaseChain: A: PDB Molecule: alkylhalidase cmls;PDBTitle: crystal structure of cmls, a flavin-dependent halogenase
11 c1pn0A_
100.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: phenol 2-monooxygenase;PDBTitle: phenol hydroxylase from trichosporon cutaneum
12 c3ihgA_
100.0
18
PDB header: flavoprotein, oxidoreductaseChain: A: PDB Molecule: rdme;PDBTitle: crystal structure of a ternary complex of aklavinone-11 hydroxylase2 with fad and aklavinone
13 c2r0gB_
100.0
22
PDB header: oxidoreductaseChain: B: PDB Molecule: rebc;PDBTitle: chromopyrrolic acid-soaked rebc with bound 7-carboxy-k252c
14 c3nixF_
100.0
15
PDB header: oxidoreductaseChain: F: PDB Molecule: flavoprotein/dehydrogenase;PDBTitle: crystal structure of flavoprotein/dehydrogenase from cytophaga2 hutchinsonii. northeast structural genomics consortium target chr43.
15 c5fn0C_
100.0
18
PDB header: oxidoreductaseChain: C: PDB Molecule: kynurenine 3-monooxygenase;PDBTitle: crystal structure of pseudomonas fluorescens kynurenine-3-2 monooxygenase (kmo) in complex with gsk180
16 c3e1tA_
100.0
16
PDB header: flavoproteinChain: A: PDB Molecule: halogenase;PDBTitle: structure and action of the myxobacterial chondrochloren2 halogenase cndh, a new variant of fad-dependent halogenases
17 c5tulA_
100.0
16
PDB header: oxidoreductaseChain: A: PDB Molecule: tetracycline destructase tet(55);PDBTitle: crystal structure of tetracycline destructase tet(55)
18 c4k22A_
100.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: protein visc;PDBTitle: structure of the c-terminal truncated form of e.coli c5-hydroxylase2 ubii involved in ubiquinone (q8) biosynthesis
19 c6bznA_
100.0
16
PDB header: flavoproteinChain: A: PDB Molecule: halogenase pltm;PDBTitle: crystal structure of halogenase pltm
20 c1phhA_
100.0
20
PDB header: oxidoreductaseChain: A: PDB Molecule: p-hydroxybenzoate hydroxylase;PDBTitle: crystal structure of p-hydroxybenzoate hydroxylase complexed with its2 reaction product 3,4-dihydroxybenzoate
21 c6aioA_
not modelled
100.0
20
PDB header: flavoproteinChain: A: PDB Molecule: pnpa;PDBTitle: crystal structure of p-nitrophenol 4-monooxygenase pnpa from2 pseudomonas putida dll-e4
22 c5dbjA_
not modelled
100.0
17
PDB header: flavoproteinChain: A: PDB Molecule: fadh2-dependent halogenase plta;PDBTitle: crystal structure of halogenase plta
23 c3gmbB_
not modelled
100.0
20
PDB header: oxidoreductaseChain: B: PDB Molecule: 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase;PDBTitle: crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid2 oxygenase
24 c4n9xA_
not modelled
100.0
20
PDB header: oxidoreductaseChain: A: PDB Molecule: putative monooxygenase;PDBTitle: crystal structure of the octaprenyl-methyl-methoxy-benzq molecule from2 erwina carotovora subsp. atroseptica strain scri 1043 / atcc baa-672,3 northeast structural genomics consortium (nesg) target ewr161
25 c3allA_
not modelled
100.0
20
PDB header: oxidoreductaseChain: A: PDB Molecule: 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase;PDBTitle: crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid2 oxygenase, mutant y270a
26 c5tukC_
not modelled
100.0
15
PDB header: oxidoreductaseChain: C: PDB Molecule: tetracycline destructase tet(51);PDBTitle: crystal structure of tetracycline destructase tet(51)
27 c2rgjA_
not modelled
100.0
20
PDB header: oxidoreductaseChain: A: PDB Molecule: flavin-containing monooxygenase;PDBTitle: crystal structure of flavin-containing monooxygenase phzs
28 c4bk2A_
not modelled
100.0
19
PDB header: oxidoreductaseChain: A: PDB Molecule: probable salicylate monooxygenase;PDBTitle: crystal structure of 3-hydroxybenzoate 6-hydroxylase2 uncovers lipid-assisted flavoprotein strategy for3 regioselective aromatic hydroxylation: q301e mutant
29 c5bulA_
not modelled
100.0
15
PDB header: hydrolaseChain: A: PDB Molecule: flavin-dependent halogenase triple mutant;PDBTitle: structure of flavin-dependent brominase bmp2 triple mutant y302s f306v2 a345w
30 c5bukA_
not modelled
100.0
17
PDB header: oxidoreductaseChain: A: PDB Molecule: fadh2-dependent halogenase;PDBTitle: structure of flavin-dependent chlorinase mpy16
31 c5wgyA_
not modelled
100.0
14
PDB header: oxidoreductase/oxidoreductase inhibitorChain: A: PDB Molecule: flavin-dependent halogenase;PDBTitle: crystal structure of mala' c112s/c128s, malbrancheamide b complex
32 c5tueB_
not modelled
100.0
15
PDB header: oxidoreductaseChain: B: PDB Molecule: tetracycline destructase tet(50);PDBTitle: crystal structure of tetracycline destructase tet(50)
33 c4cy8A_
not modelled
100.0
19
PDB header: oxidoreductaseChain: A: PDB Molecule: 2-hydroxybiphenyl 3-monooxygenase;PDBTitle: 2-hydroxybiphenyl 3-monooxygenase (hbpa) in complex with fad
34 c5x68B_
not modelled
100.0
17
PDB header: oxidoreductaseChain: B: PDB Molecule: kynurenine 3-monooxygenase;PDBTitle: crystal structure of human kmo
35 c6bz5B_
not modelled
100.0
20
PDB header: oxidoreductaseChain: B: PDB Molecule: salicylate hydroxylase;PDBTitle: structure and mechanism of salicylate hydroxylase from pseudomonas2 putida g7
36 c2x3nA_
not modelled
100.0
20
PDB header: oxidoreductaseChain: A: PDB Molecule: probable fad-dependent monooxygenase;PDBTitle: crystal structure of pqsl, a probable fad-dependent monooxygenase from2 pseudomonas aeruginosa
37 c3rp7A_
not modelled
100.0
16
PDB header: oxidoreductaseChain: A: PDB Molecule: flavoprotein monooxygenase;PDBTitle: crystal structure of klebsiella pneumoniae hpxo complexed with fad and2 uric acid
38 c5evyX_
not modelled
100.0
19
PDB header: oxidoreductaseChain: X: PDB Molecule: salicylate hydroxylase;PDBTitle: salicylate hydroxylase substrate complex
39 c5eowA_
not modelled
100.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: 6-hydroxynicotinate 3-monooxygenase;PDBTitle: crystal structure of 6-hydroxynicotinic acid 3-monooxygenase from2 pseudomonas putida kt2440
40 c4j33B_
not modelled
100.0
16
PDB header: oxidoreductaseChain: B: PDB Molecule: kynurenine 3-monooxygenase;PDBTitle: crystal structure of kynurenine 3-monooxygenase (kmo-394)
41 c3c4aA_
not modelled
100.0
13
PDB header: oxidoreductaseChain: A: PDB Molecule: probable tryptophan hydroxylase viod;PDBTitle: crystal structure of viod hydroxylase in complex with fad from2 chromobacterium violaceum. northeast structural genomics consortium3 target cvr158
42 d1k0ia1
not modelled
100.0
24
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain
43 c3cgvA_
not modelled
100.0
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: geranylgeranyl reductase related protein;PDBTitle: crystal structure of geranylgeranyl bacteriochlorophyll reductase-like2 fixc homolog (np_393992.1) from thermoplasma acidophilum at 1.60 a3 resolution
44 c3atrA_
not modelled
100.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: conserved archaeal protein;PDBTitle: geranylgeranyl reductase (ggr) from sulfolobus acidocaldarius co-2 crystallized with its ligand
45 c2vouA_
not modelled
100.0
17
PDB header: oxidoreductaseChain: A: PDB Molecule: 2,6-dihydroxypyridine hydroxylase;PDBTitle: structure of 2,6-dihydroxypyridine-3-hydroxylase from2 arthrobacter nicotinovorans
46 d3c96a1
not modelled
100.0
25
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain
47 c2bryA_
not modelled
100.0
17
PDB header: transportChain: A: PDB Molecule: nedd9 interacting protein with calponin homologyPDBTitle: crystal structure of the native monooxygenase domain of2 mical at 1.45 a resolution
48 d1pn0a1
not modelled
100.0
13
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain
49 c2xdoC_
not modelled
100.0
17
PDB header: oxidoreductaseChain: C: PDB Molecule: tetx2 protein;PDBTitle: structure of the tetracycline degrading monooxygenase tetx2 from2 bacteroides thetaiotaomicron
50 c4hb9A_
not modelled
100.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: similarities with probable monooxygenase;PDBTitle: crystal structure of a putative fad containing monooxygenase from2 photorhabdus luminescens subsp. laumondii tto1 (target psi-012791)
51 c5uaoA_
not modelled
100.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: tryptophane-5-halogenase;PDBTitle: crystal structure of mibh, a lathipeptide tryptophan 5-halogenase
52 c4txkA_
not modelled
100.0
16
PDB header: oxidoreductaseChain: A: PDB Molecule: protein-methionine sulfoxide oxidase mical1;PDBTitle: construct of mical-1 containing the monooxygenase and calponin2 homology domains
53 c2ardA_
not modelled
100.0
16
PDB header: biosynthetic proteinChain: A: PDB Molecule: tryptophan halogenase prna;PDBTitle: the structure of tryptophan 7-halogenase (prna) suggests a mechanism2 for regioselective chlorination
54 c5hy5A_
not modelled
100.0
17
PDB header: oxidoreductaseChain: A: PDB Molecule: tryptophan 6-halogenase;PDBTitle: crystal structure of a tryptophan 6-halogenase (stth) from2 streptomyces toxytricini
55 c3ihmB_
not modelled
100.0
16
PDB header: oxidoreductaseChain: B: PDB Molecule: styrene monooxygenase a;PDBTitle: structure of the oxygenase component of a pseudomonas styrene2 monooxygenase
56 c6frlA_
not modelled
100.0
16
PDB header: flavoproteinChain: A: PDB Molecule: tryptophan halogenase superfamily;PDBTitle: brvh, a flavin-dependent halogenase from brevundimonas sp. bal3
57 d2voua1
not modelled
100.0
25
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain
58 c2weuD_
not modelled
100.0
21
PDB header: antifungal proteinChain: D: PDB Molecule: tryptophan 5-halogenase;PDBTitle: crystal structure of tryptophan 5-halogenase (pyrh) complex2 with substrate tryptophan
59 c6ib5B_
not modelled
100.0
17
PDB header: flavoproteinChain: B: PDB Molecule: tryptophan 6-halogenase;PDBTitle: mutant of flavin-dependent tryptophan halogenase thal with altered2 regioselectivity (thal-rebh5)
60 c2e4gB_
not modelled
100.0
18
PDB header: biosynthetic protein, flavoproteinChain: B: PDB Molecule: tryptophan halogenase;PDBTitle: rebh with bound l-trp
61 c2pyxA_
not modelled
100.0
19
PDB header: biosynthetic proteinChain: A: PDB Molecule: tryptophan halogenase;PDBTitle: crystal structure of tryptophan halogenase (yp_750003.1) from2 shewanella frigidimarina ncimb 400 at 1.50 a resolution
62 d2gmha1
not modelled
100.0
17
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain
63 c2gmhA_
not modelled
100.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: electron transfer flavoprotein-ubiquinonePDBTitle: structure of porcine electron transfer flavoprotein-2 ubiquinone oxidoreductase in complexed with ubiquinone
64 c3nrnA_
not modelled
99.9
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein pf1083;PDBTitle: crystal structure of pf1083 protein from pyrococcus furiosus,2 northeast structural genomics consortium target pfr223
65 c1yvvB_
not modelled
99.8
13
PDB header: oxidoreductaseChain: B: PDB Molecule: amine oxidase, flavin-containing;PDBTitle: x-ray structurure of p. syringae q888a4 oxidoreductase at2 resolution 2.5a. northeast structural genomics consortium3 target psr10.
66 c5ez7A_
not modelled
99.8
18
PDB header: oxidoreductaseChain: A: PDB Molecule: flavoenzyme pa4991;PDBTitle: crystal structure of the fad dependent oxidoreductase pa4991 from2 pseudomonas aeruginosa
67 c3da1A_
not modelled
99.8
13
PDB header: oxidoreductaseChain: A: PDB Molecule: glycerol-3-phosphate dehydrogenase;PDBTitle: x-ray structure of the glycerol-3-phosphate dehydrogenase2 from bacillus halodurans complexed with fad. northeast3 structural genomics consortium target bhr167.
68 c3qj4A_
not modelled
99.7
12
PDB header: oxidoreductaseChain: A: PDB Molecule: renalase;PDBTitle: crystal structure of human renalase (isoform 1)
69 c1y56B_
not modelled
99.7
18
PDB header: oxidoreductaseChain: B: PDB Molecule: sarcosine oxidase;PDBTitle: crystal structure of l-proline dehydrogenase from p.horikoshii
70 c3nyeA_
not modelled
99.7
18
PDB header: oxidoreductaseChain: A: PDB Molecule: d-arginine dehydrogenase;PDBTitle: crystal structure of pseudomonas aeruginosa d-arginine dehydrogenase2 in complex with imino-arginine
71 c3ka7A_
not modelled
99.7
18
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase;PDBTitle: crystal structure of an oxidoreductase from methanosarcina2 mazei. northeast structural genomics consortium target id3 mar208
72 c4p9sA_
not modelled
99.7
19
PDB header: oxidoreductaseChain: A: PDB Molecule: dimethylglycine dehydrogenase;PDBTitle: crystal structure of the mature form of rat dmgdh
73 c2r4jA_
not modelled
99.7
17
PDB header: oxidoreductaseChain: A: PDB Molecule: aerobic glycerol-3-phosphate dehydrogenase;PDBTitle: crystal structure of escherichia coli semet substituted glycerol-3-2 phosphate dehydrogenase in complex with dhap
74 c2olnA_
not modelled
99.7
16
PDB header: oxidoreductaseChain: A: PDB Molecule: nikd protein;PDBTitle: nikd, an unusual amino acid oxidase essential for2 nikkomycin biosynthesis: closed form at 1.15 a resolution
75 c6j39A_
not modelled
99.6
17
PDB header: oxidoreductase/inhibitorChain: A: PDB Molecule: fad-dependent glycine oxydase;PDBTitle: crystal structure of cmis2 with inhibitor
76 c3bhkA_
not modelled
99.6
16
PDB header: oxidoreductaseChain: A: PDB Molecule: monomeric sarcosine oxidase;PDBTitle: crystal structure of r49k mutant of monomeric sarcosine oxidase2 crystallized in phosphate as precipitant
77 d2ivda1
not modelled
99.6
26
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain
78 c3ps9A_
not modelled
99.6
18
PDB header: transferaseChain: A: PDB Molecule: trna 5-methylaminomethyl-2-thiouridine biosynthesisPDBTitle: crystal structure of mnmc from e. coli
79 c3dmeB_
not modelled
99.6
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: conserved exported protein;PDBTitle: crystal structure of conserved exported protein from bordetella2 pertussis. northeast structural genomics target ber141
80 c1pj6A_
not modelled
99.6
20
PDB header: oxidoreductaseChain: A: PDB Molecule: n,n-dimethylglycine oxidase;PDBTitle: crystal structure of dimethylglycine oxidase of arthrobacter2 globiformis in complex with folic acid
81 c5ttkB_
not modelled
99.6
14
PDB header: oxidoreductaseChain: B: PDB Molecule: amine oxidase;PDBTitle: crystal structure of selenomethionine-incorporated nicotine2 oxidoreductase from pseudomonas putida
82 c5g3sB_
not modelled
99.6
13
PDB header: oxidoreductaseChain: B: PDB Molecule: l-tryptophan oxidase vioa;PDBTitle: the structure of the l-tryptophan oxidase vioa from chromobacterium2 violaceum - samarium derivative
83 c5twcA_
not modelled
99.6
14
PDB header: oxidoreductaseChain: A: PDB Molecule: ferredoxin--nadp reductase;PDBTitle: oxidoreductase iruo in the oxidized form
84 c2vvlD_
not modelled
99.6
13
PDB header: oxidoreductaseChain: D: PDB Molecule: monoamine oxidase n;PDBTitle: the structure of mao-n-d3, a variant of monoamine oxidase2 from aspergillus niger.
85 c3axbA_
not modelled
99.6
15
PDB header: oxidoreductaseChain: A: PDB Molecule: putative oxidoreductase;PDBTitle: structure of a dye-linked l-proline dehydrogenase from the aerobic2 hyperthermophilic archaeon, aeropyrum pernix
86 c4x9mA_
not modelled
99.6
14
PDB header: oxidoreductaseChain: A: PDB Molecule: l-alpha-glycerophosphate oxidase;PDBTitle: oxidized l-alpha-glycerophosphate oxidase from mycoplasma pneumoniae2 with fad bound
87 c2ivdA_
not modelled
99.6
22
PDB header: oxidoreductaseChain: A: PDB Molecule: protoporphyrinogen oxidase;PDBTitle: structure of protoporphyrinogen oxidase from myxococcus2 xanthus with acifluorfen
88 d2gqfa1
not modelled
99.6
21
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: HI0933 N-terminal domain-like
89 c3i6dA_
not modelled
99.6
19
PDB header: oxidoreductaseChain: A: PDB Molecule: protoporphyrinogen oxidase;PDBTitle: crystal structure of ppo from bacillus subtilis with af
90 c6cr0A_
not modelled
99.6
12
PDB header: oxidoreductaseChain: A: PDB Molecule: (s)-6-hydroxynicotine oxidase;PDBTitle: 1.55 a resolution structure of (s)-6-hydroxynicotine oxidase from2 shinella hzn7
91 c3f8rD_
not modelled
99.6
19
PDB header: oxidoreductaseChain: D: PDB Molecule: thioredoxin reductase (trxb-3);PDBTitle: crystal structure of sulfolobus solfataricus thioredoxin2 reductase b3 in complex with two nadp molecules
92 c3pvcA_
not modelled
99.6
20
PDB header: oxidoreductase, transferaseChain: A: PDB Molecule: trna 5-methylaminomethyl-2-thiouridine biosynthesisPDBTitle: crystal structure of apo mnmc from yersinia pestis
93 c5mogB_
not modelled
99.6
15
PDB header: oxidoreductaseChain: B: PDB Molecule: phytoene dehydrogenase, chloroplastic/chromoplastic;PDBTitle: oryza sativa phytoene desaturase inhibited by norflurazon
94 c2gahB_
not modelled
99.6
18
PDB header: oxidoreductaseChain: B: PDB Molecule: heterotetrameric sarcosine oxidase beta-subunit;PDBTitle: heterotetrameric sarcosine: structure of a diflavin2 metaloenzyme at 1.85 a resolution
95 c4xwzA_
not modelled
99.6
16
PDB header: oxidoreductaseChain: A: PDB Molecule: fructosyl amine:oxygen oxidoreductase;PDBTitle: the crystal structure of fructosyl amine: oxygen oxidoreductase2 (amadoriase i) from aspergillus fumigatus in complex with the3 substrate fructosyl lysine
96 c2rgoA_
not modelled
99.6
18
PDB header: oxidoreductaseChain: A: PDB Molecule: alpha-glycerophosphate oxidase;PDBTitle: structure of alpha-glycerophosphate oxidase from2 streptococcus sp.: a template for the mitochondrial alpha-3 glycerophosphate dehydrogenase
97 d1b5qa1
not modelled
99.6
19
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain
98 d1kf6a2
not modelled
99.6
17
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain
99 c6garB_
not modelled
99.5
13
PDB header: oxidoreductaseChain: B: PDB Molecule: ferredoxin--nadp reductase;PDBTitle: crystal structure of oxidised ferredoxin/flavodoxin nadp+2 oxidoreductase 1 (fnr1) from bacillus cereus
100 c4y4nE_
not modelled
99.5
25
PDB header: biosynthetic proteinChain: E: PDB Molecule: putative ribose 1,5-bisphosphate isomerase;PDBTitle: thiazole synthase thi4 from methanococcus igneus
101 c1ryiB_
not modelled
99.5
15
PDB header: oxidoreductaseChain: B: PDB Molecule: glycine oxidase;PDBTitle: structure of glycine oxidase with bound inhibitor glycolate
102 d1reoa1
not modelled
99.5
17
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain
103 c3jskN_
not modelled
99.5
20
PDB header: biosynthetic proteinChain: N: PDB Molecule: cypbp37 protein;PDBTitle: thiazole synthase from neurospora crassa
104 c2rghA_
not modelled
99.5
18
PDB header: oxidoreductaseChain: A: PDB Molecule: alpha-glycerophosphate oxidase;PDBTitle: structure of alpha-glycerophosphate oxidase from2 streptococcus sp.: a template for the mitochondrial alpha-3 glycerophosphate dehydrogenase
105 d1neka2
not modelled
99.5
13
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain
106 c4dgkA_
not modelled
99.5
16
PDB header: oxidoreductaseChain: A: PDB Molecule: phytoene dehydrogenase;PDBTitle: crystal structure of phytoene desaturase crti from pantoea ananatis
107 c1c0iA_
not modelled
99.5
14
PDB header: oxidoreductaseChain: A: PDB Molecule: d-amino acid oxidase;PDBTitle: crystal structure of d-amino acid oxidase in complex with two2 anthranylate molecules
108 c4repA_
not modelled
99.5
22
PDB header: oxidoreductase, flavoproteinChain: A: PDB Molecule: gamma-carotene desaturase;PDBTitle: crystal structure of gamma-carotenoid desaturase
109 c1s3bB_
not modelled
99.5
16
PDB header: oxidoreductaseChain: B: PDB Molecule: amine oxidase [flavin-containing] b;PDBTitle: crystal structure of maob in complex with n-methyl-n-2 propargyl-1(r)-aminoindan
110 c3ab1B_
not modelled
99.5
17
PDB header: oxidoreductaseChain: B: PDB Molecule: ferredoxin--nadp reductase;PDBTitle: crystal structure of ferredoxin nadp+ oxidoreductase
111 d2bs2a2
not modelled
99.5
19
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain
112 c4i58A_
not modelled
99.5
19
PDB header: oxidoreductaseChain: A: PDB Molecule: cyclohexylamine oxidase;PDBTitle: cyclohexylamine oxidase from brevibacterium oxydans ih-35a
113 c2zxiC_
not modelled
99.5
19
PDB header: fad-binding proteinChain: C: PDB Molecule: trna uridine 5-carboxymethylaminomethyl modification enzymePDBTitle: structure of aquifex aeolicus gida in the form ii crystal
114 c1f8sA_
not modelled
99.5
15
PDB header: oxidoreductaseChain: A: PDB Molecule: l-amino acid oxidase;PDBTitle: crystal structure of l-amino acid oxidase from calloselasma2 rhodostoma, complexed with three molecules of o-aminobenzoate.
115 c3djeA_
not modelled
99.5
16
PDB header: oxidoreductaseChain: A: PDB Molecule: fructosyl amine: oxygen oxidoreductase;PDBTitle: crystal structure of the deglycating enzyme fructosamine2 oxidase from aspergillus fumigatus (amadoriase ii) in3 complex with fsa
116 d1jnra2
not modelled
99.5
17
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain
117 c5fjnB_
not modelled
99.5
20
PDB header: hydrolaseChain: B: PDB Molecule: l-amino acid deaminase;PDBTitle: structure of l-amino acid deaminase from proteus myxofaciens2 in complex with anthranilate
118 d1qo8a2
not modelled
99.5
18
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain
119 c4yshA_
not modelled
99.4
18
PDB header: oxidoreductaseChain: A: PDB Molecule: glycine oxidase;PDBTitle: crystal structure of glycine oxidase from geobacillus kaustophilus
120 c3cp2A_
not modelled
99.4
21
PDB header: oxidoreductaseChain: A: PDB Molecule: trna uridine 5-carboxymethylaminomethylPDBTitle: crystal structure of gida from e. coli