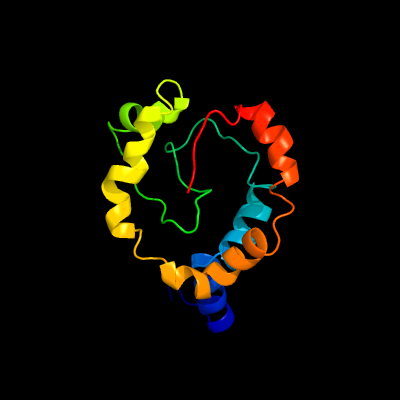
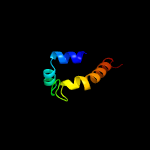
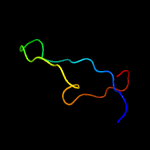
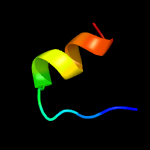
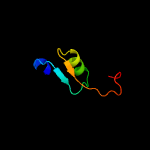
1 c2k3mA_
100.0
98
PDB header: membrane proteinChain: A: PDB Molecule: rv1761c;PDBTitle: rv1761c
2 c3n6yA_
37.9
26
PDB header: unknown functionChain: A: PDB Molecule: immunoglobulin-like protein;PDBTitle: crystal structure of an immunoglobulin-like protein (pa1606) from2 pseudomonas aeruginosa at 1.50 a resolution
3 d1fn9a_
16.8
34
Fold: Outer capsid protein sigma 3Superfamily: Outer capsid protein sigma 3Family: Outer capsid protein sigma 3
4 c1g0vB_
15.0
30
PDB header: hydrolase/hydrolase inhibitorChain: B: PDB Molecule: protease a inhibitor 3;PDBTitle: the structure of proteinase a complexed with a ia3 mutant, mvv
5 c2xppB_
13.1
50
PDB header: transcriptionChain: B: PDB Molecule: chromatin structure modulator;PDBTitle: crystal structure of a spt6-iws1(spn1) complex from2 encephalitozoon cuniculi, form iii
6 d1zs4a1
12.4
31
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Bacteriophage CII protein
7 c1u0lB_
12.3
30
PDB header: hydrolaseChain: B: PDB Molecule: probable gtpase engc;PDBTitle: crystal structure of yjeq from thermotoga maritima
8 c4bjjA_
12.0
27
PDB header: transcriptionChain: A: PDB Molecule: transcription factor tau subunit sfc1;PDBTitle: sfc1-sfc7 dimerization module
9 c4phtC_
12.0
23
PDB header: protein transportChain: C: PDB Molecule: general secretory pathway protein e;PDBTitle: atpase gspe in complex with the cytoplasmic domain of gspl from the2 vibrio vulnificus type ii secretion system
10 c1mqrA_
11.7
33
PDB header: hydrolaseChain: A: PDB Molecule: alpha-d-glucuronidase;PDBTitle: the crystal structure of alpha-d-glucuronidase (e386q) from bacillus2 stearothermophilus t-6
11 d1h41a1
11.5
26
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: alpha-D-glucuronidase/Hyaluronidase catalytic domain
12 c5aa6F_
11.3
11
PDB header: oxidoreductaseChain: F: PDB Molecule: vanadium-dependent bromoperoxidase 2;PDBTitle: homohexameric structure of the second vanadate-dependent2 bromoperoxidase (anii) from ascophyllum nodosum
13 d1l8na1
11.1
33
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: alpha-D-glucuronidase/Hyaluronidase catalytic domain
14 d1yq2a3
10.6
25
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: beta-Galactosidase/glucuronidase, N-terminal domain
15 d1t3va_
10.2
10
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like
16 c5zfqA_
9.6
18
PDB header: transport proteinChain: A: PDB Molecule: twitching motility pilus retraction protein;PDBTitle: crystal structure of pilt-4, a retraction atpase motor of type iv2 pilus , from geobacter sulfurreducens
17 d1p9ra_
9.4
23
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)
18 d1t9ha1
9.2
44
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
19 d1n6za_
9.1
45
Fold: Hypothetical protein Yml108wSuperfamily: Hypothetical protein Yml108wFamily: Hypothetical protein Yml108w
20 c5oqml_
9.0
18
PDB header: transcriptionChain: L: PDB Molecule: dna-directed rna polymerases i, ii, and iii subunit rpabc4;PDBTitle: structure of yeast transcription pre-initiation complex with tfiih and2 core mediator
21 d1jala2
not modelled
8.9
47
Fold: beta-Grasp (ubiquitin-like)Superfamily: TGS-likeFamily: G domain-linked domain
22 d1knxa1
not modelled
8.4
33
Fold: MurF and HprK N-domain-likeSuperfamily: HprK N-terminal domain-likeFamily: HPr kinase/phoshatase HprK N-terminal domain
23 c4kssC_
not modelled
8.3
29
PDB header: protein transportChain: C: PDB Molecule: type ii secretion system protein e, hemolysin-coregulatedPDBTitle: crystal structure of vibrio cholerae atpase gspse hexamer
24 d1ni3a2
not modelled
8.1
40
Fold: beta-Grasp (ubiquitin-like)Superfamily: TGS-likeFamily: G domain-linked domain
25 d1o13a_
not modelled
8.0
10
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like
26 d1xkpa1
not modelled
7.9
20
Fold: Type III secretion system domainSuperfamily: Type III secretion system domainFamily: LcrE-like
27 c2xpoD_
not modelled
7.6
55
PDB header: transcriptionChain: D: PDB Molecule: chromatin structure modulator;PDBTitle: crystal structure of a spt6-iws1(spn1) complex from2 encephalitozoon cuniculi, form ii
28 c2xpoB_
not modelled
7.4
55
PDB header: transcriptionChain: B: PDB Molecule: chromatin structure modulator;PDBTitle: crystal structure of a spt6-iws1(spn1) complex from2 encephalitozoon cuniculi, form ii
29 d2al6a3
not modelled
7.4
38
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: First domain of FERM
30 c1gqkB_
not modelled
7.1
26
PDB header: hydrolaseChain: B: PDB Molecule: alpha-d-glucuronidase;PDBTitle: structure of pseudomonas cellulosa alpha-d-glucuronidase complexed2 with glucuronic acid
31 c2xpnB_
not modelled
6.8
55
PDB header: transcriptionChain: B: PDB Molecule: chromatin structure modulator;PDBTitle: crystal structure of a spt6-iws1(spn1) complex from2 encephalitozoon cuniculi, form i
32 c2eouA_
not modelled
6.5
40
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 473;PDBTitle: solution structure of the c2h2 type zinc finger (region 370-2 400) of human zinc finger protein 473
33 d1ddba_
not modelled
6.5
23
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death
34 c3jvvA_
not modelled
6.5
35
PDB header: atp binding proteinChain: A: PDB Molecule: twitching mobility protein;PDBTitle: crystal structure of p. aeruginosa pilt with bound amp-pcp
35 d2c0sa1
not modelled
6.1
32
Fold: ROP-likeSuperfamily: BAS1536-likeFamily: BAS1536-like
36 c2npbA_
not modelled
5.9
24
PDB header: oxidoreductaseChain: A: PDB Molecule: selenoprotein w;PDBTitle: nmr solution structure of mouse selw
37 c5fl3A_
not modelled
5.9
29
PDB header: transport proteinChain: A: PDB Molecule: pili retraction protein pilt;PDBTitle: pilt2 from thermus thermophilus
38 d2bzba1
not modelled
5.8
56
Fold: ROP-likeSuperfamily: BAS1536-likeFamily: BAS1536-like
39 c1knxF_
not modelled
5.7
33
PDB header: transferase/hydrolaseChain: F: PDB Molecule: probable hpr(ser) kinase/phosphatase;PDBTitle: hpr kinase/phosphatase from mycoplasma pneumoniae
40 c2yx6C_
not modelled
5.7
12
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein ph0822;PDBTitle: crystal structure of ph0822
41 c3ebwA_
not modelled
5.5
29
PDB header: allergenChain: A: PDB Molecule: per a 4 allergen;PDBTitle: crystal structure of major allergens, per a 4 from cockroaches
42 c5tshF_
not modelled
5.4
47
PDB header: atp-binding proteinChain: F: PDB Molecule: type iv pilus biogenesis atpase pilb;PDBTitle: pilb from geobacter metallireducens bound to amp-pnp
43 d1ys7a1
not modelled
5.3
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: PhoB-like
44 d2j8xa1
not modelled
5.2
25
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Uracil-DNA glycosylase
45 c4lrvL_
not modelled
5.2
48
PDB header: dna binding proteinChain: L: PDB Molecule: dna sulfur modification protein dnde;PDBTitle: crystal structure of dnde from escherichia coli b7a involved in dna2 phosphorothioation modification
46 c5bncB_
not modelled
5.1
17
PDB header: heme binding proteinChain: B: PDB Molecule: heme binding protein msmeg_6519;PDBTitle: structure of heme binding protein msmeg_6519 from mycobacterium2 smegmatis
47 c6gefB_
not modelled
5.1
35
PDB header: hydrolaseChain: B: PDB Molecule: type iv secretion system protein dotb;PDBTitle: x-ray structure of the yersinia pseudotuberculosis atpase dotb
48 c3graA_
not modelled
5.1
23
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, arac family;PDBTitle: crystal structure of arac family transcriptional regulator from2 pseudomonas putida
49 d2dexx3
not modelled
5.0
37
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Peptidylarginine deiminase Pad4, catalytic C-terminal domain