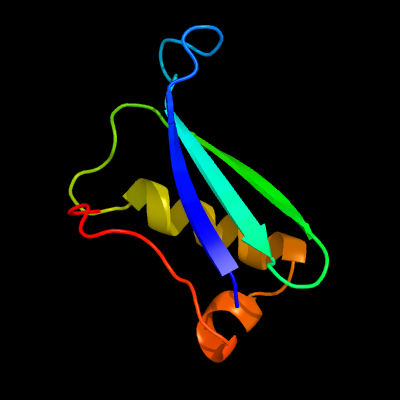
1 c1c0mA_
99.1
20
PDB header: transferaseChain: A: PDB Molecule: protein (integrase);PDBTitle: crystal structure of rsv two-domain integrase
2 d1asua_
99.0
20
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain
3 d1cxqa_
99.0
20
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain
4 d1c0ma2
99.0
20
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain
5 c5ejkG_
98.9
20
PDB header: transferase/dnaChain: G: PDB Molecule: gag-pro-pol polyprotein;PDBTitle: crystal structure of the rous sarcoma virus intasome
6 d1bcoa2
98.9
26
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: mu transposase, core domain
7 c5cz1B_
98.7
15
PDB header: hydrolaseChain: B: PDB Molecule: integrase;PDBTitle: crystal structure of the catalytic core domain of mmtv integrase
8 c4mq3A_
98.7
21
PDB header: viral proteinChain: A: PDB Molecule: integrase;PDBTitle: the 1.1 angstrom structure of catalytic core domain of fiv integrase
9 c3nf9A_
98.6
18
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: integrase;PDBTitle: structural basis for a new mechanism of inhibition of hiv integrase2 identified by fragment screening and structure based design
10 c4fcyA_
98.5
24
PDB header: dna binding protein/dnaChain: A: PDB Molecule: transposase;PDBTitle: crystal structure of the bacteriophage mu transpososome
11 c3jcaE_
98.5
17
PDB header: viral proteinChain: E: PDB Molecule: integrase;PDBTitle: core model of the mouse mammary tumor virus intasome
12 d1c6va_
98.4
15
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain
13 c5u1cD_
98.3
18
PDB header: viral proteinChain: D: PDB Molecule: hiv-1 integrase, sso7d chimera;PDBTitle: structure of tetrameric hiv-1 strand transfer complex intasome
14 d1exqa_
98.3
18
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain
15 c3kksB_
98.3
20
PDB header: dna binding proteinChain: B: PDB Molecule: integrase;PDBTitle: crystal structure of catalytic core domain of biv integrase in crystal2 form ii
16 c1ex4A_
98.3
18
PDB header: viral proteinChain: A: PDB Molecule: integrase;PDBTitle: hiv-1 integrase catalytic core and c-terminal domain
17 c1bcoA_
98.1
23
PDB header: transposaseChain: A: PDB Molecule: bacteriophage mu transposase;PDBTitle: bacteriophage mu transposase core domain
18 c3f9kV_
98.1
13
PDB header: viral protein, recombinationChain: V: PDB Molecule: integrase;PDBTitle: two domain fragment of hiv-2 integrase in complex with ledgf ibd
19 d1hyva_
97.9
18
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain
20 c5u1cA_
97.9
18
PDB header: viral proteinChain: A: PDB Molecule: hiv-1 integrase, sso7d chimera;PDBTitle: structure of tetrameric hiv-1 strand transfer complex intasome
21 c3dlrA_
not modelled
97.5
23
PDB header: transferaseChain: A: PDB Molecule: integrase;PDBTitle: crystal structure of the catalytic core domain from pfv integrase
22 c1k6yB_
not modelled
97.4
18
PDB header: transferaseChain: B: PDB Molecule: integrase;PDBTitle: crystal structure of a two-domain fragment of hiv-1 integrase
23 c3hpgC_
not modelled
97.3
15
PDB header: transferaseChain: C: PDB Molecule: integrase;PDBTitle: visna virus integrase (residues 1-219) in complex with ledgf2 ibd: examples of open integrase dimer-dimer interfaces
24 c5m0rF_
not modelled
97.0
18
PDB header: hydrolaseChain: F: PDB Molecule: integrase;PDBTitle: cryo-em reconstruction of the maedi-visna virus (mvv) strand transfer2 complex
25 c3l2tB_
not modelled
96.2
23
PDB header: recombination/dnaChain: B: PDB Molecule: integrase;PDBTitle: crystal structure of the prototype foamy virus (pfv) intasome in2 complex with magnesium and mk0518 (raltegravir)
26 c5xnwA_
not modelled
27.2
26
PDB header: lyaseChain: A: PDB Molecule: adenylate cyclase exoy;PDBTitle: crystal structure of exoy, a unique nucleotidyl cyclase toxin from2 pseudomonas aeruginosa
27 d1s99a_
not modelled
22.5
24
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: Putative thiamin/HMP-binding protein YkoF
28 c2xk0A_
not modelled
21.1
15
PDB header: transcriptionChain: A: PDB Molecule: polycomb protein pcl;PDBTitle: solution structure of the tudor domain from drosophila2 polycomblike (pcl)
29 c3ks7D_
not modelled
18.7
29
PDB header: hydrolaseChain: D: PDB Molecule: putative putative pngase f;PDBTitle: crystal structure of putative peptide:n-glycosidase f (pngase f)2 (yp_210507.1) from bacteroides fragilis nctc 9343 at 2.30 a3 resolution
30 d1d6za3
not modelled
17.9
28
Fold: Cystatin-likeSuperfamily: Amine oxidase N-terminal regionFamily: Amine oxidase N-terminal region
31 c4f48A_
not modelled
15.5
12
PDB header: signaling proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: the x-ray structural of fimxeal-c-di-gmp-pilz complexes from2 xanthomonas campestris
32 c2e4jA_
not modelled
15.2
13
PDB header: isomeraseChain: A: PDB Molecule: prostaglandin-h2 d-isomerase;PDBTitle: solution structure of mouse lipocalin-type prostaglandin d2 synthase
33 c5ho0A_
not modelled
13.6
21
PDB header: hydrolaseChain: A: PDB Molecule: extracellular arabinanase;PDBTitle: crystal structure of abna (closed conformation), a gh43 extracellular2 arabinanase from geobacillus stearothermophilus
34 c3pz8A_
not modelled
13.2
25
PDB header: signaling proteinChain: A: PDB Molecule: segment polarity protein dishevelled homolog dvl-1;PDBTitle: crystal structure of dvl1-dix(y17d) mutant
35 d2oqea3
not modelled
13.1
15
Fold: Cystatin-likeSuperfamily: Amine oxidase N-terminal regionFamily: Amine oxidase N-terminal region
36 c3cqrB_
not modelled
12.1
9
PDB header: oxidoreductaseChain: B: PDB Molecule: violaxanthin de-epoxidase, chloroplast;PDBTitle: crystal structure of the lipocalin domain of violaxanthin de-epoxidase2 (vde) at ph5
37 c4b6aI_
not modelled
11.8
14
PDB header: ribosomeChain: I: PDB Molecule: 60s ribosomal protein l10;PDBTitle: cryo-em structure of the 60s ribosomal subunit in complex2 with arx1 and rei1
38 c3kzpA_
not modelled
11.7
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative diguanylate cyclase/phosphodiesterase;PDBTitle: crystal structure of putative diguanylate cyclase/phosphodiesterase2 from listaria monocytigenes
39 d1w6ga3
not modelled
10.9
13
Fold: Cystatin-likeSuperfamily: Amine oxidase N-terminal regionFamily: Amine oxidase N-terminal region
40 c3lv4B_
not modelled
10.9
21
PDB header: hydrolaseChain: B: PDB Molecule: glycoside hydrolase yxia;PDBTitle: crystal structure of the glycoside hydrolase, family 43 yxia protein2 from bacillus licheniformis. northeast structural genomics consortium3 target bir14.
41 d2c9aa1
not modelled
10.5
6
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains
42 c4bwvB_
not modelled
9.3
9
PDB header: oxidoreductaseChain: B: PDB Molecule: phosphoadenosine-phosphosulphate reductase;PDBTitle: structure of adenosine 5-prime-phosphosulfate reductase apr-b from2 physcomitrella patens
43 d2basa1
not modelled
8.9
20
Fold: TIM beta/alpha-barrelSuperfamily: EAL domain-likeFamily: EAL domain
44 c2m0oA_
not modelled
8.6
21
PDB header: peptide binding proteinChain: A: PDB Molecule: phd finger protein 1;PDBTitle: the solution structure of human phf1 in complex with h3k36me3
45 c3hvbB_
not modelled
8.5
9
PDB header: hydrolaseChain: B: PDB Molecule: protein fimx;PDBTitle: crystal structure of the dual-domain ggdef-eal module of fimx from2 pseudomonas aeruginosa
46 c1h2iG_
not modelled
8.3
19
PDB header: dna binding proteinChain: G: PDB Molecule: dna repair protein rad52 homolog;PDBTitle: human rad52 protein, n-terminal domain
47 c4p37A_
not modelled
8.1
42
PDB header: transferaseChain: A: PDB Molecule: putative poly(a) polymerase catalytic subunit;PDBTitle: crystal structure of the megavirus polyadenylate synthase
48 c4q6jB_
not modelled
7.9
22
PDB header: unknown functionChain: B: PDB Molecule: lmo0131 protein;PDBTitle: crystal structure of eal domain protein from listeria monocytogenes2 egd-e
49 d1d6za2
not modelled
7.8
15
Fold: Cystatin-likeSuperfamily: Amine oxidase N-terminal regionFamily: Amine oxidase N-terminal region
50 c4a1aH_
not modelled
7.6
11
PDB header: ribosomeChain: H: PDB Molecule: 60s ribosomal protein l10;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 3.
51 c6n2oB_
not modelled
7.5
14
PDB header: oxidoreductaseChain: B: PDB Molecule: pyruvate ferredoxin/flavodoxin oxidoreductase, betaPDBTitle: 2-oxoglutarate:ferredoxin oxidoreductase from magnetococcus marinus2 with 2-oxoglutarate, coenzyme a and succinyl-coa bound
52 d1ebda3
not modelled
7.2
10
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domainFamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domain
53 d1kn0a_
not modelled
7.1
14
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: The homologous-pairing domain of Rad52 recombinase
54 c3s83A_
not modelled
6.3
9
PDB header: signaling proteinChain: A: PDB Molecule: ggdef family protein;PDBTitle: eal domain of phosphodiesterase pdea
55 c2k23A_
not modelled
5.9
12
PDB header: transport proteinChain: A: PDB Molecule: lipocalin 2;PDBTitle: solution structure analysis of the rlcn2
56 c3pz7A_
not modelled
5.8
25
PDB header: signaling proteinChain: A: PDB Molecule: dixin;PDBTitle: crystal structure of ccd1-dix domain
57 d1gesa3
not modelled
5.6
19
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domainFamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domain
58 c3zf7K_
not modelled
5.5
6
PDB header: ribosomeChain: K: PDB Molecule: 60s ribosomal protein l10, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
59 c3l2uA_
not modelled
5.4
22
PDB header: recombination/dnaChain: A: PDB Molecule: integrase;PDBTitle: crystal structure of the prototype foamy virus (pfv)2 intasome in complex with magnesium and gs91373 (elvitegravir)
60 c4lykB_
not modelled
5.4
13
PDB header: hydrolaseChain: B: PDB Molecule: cyclic di-gmp phosphodiesterase yaha;PDBTitle: crystal structure of the eal domain of c-di-gmp specific2 phosphodiesterase yaha in complex with activating cofactor mg++
61 c5swcE_
not modelled
5.4
11
PDB header: lyaseChain: E: PDB Molecule: carbonic anhydrase;PDBTitle: the structure of the beta-carbonic anhydrase ccaa
62 d3grsa3
not modelled
5.3
19
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domainFamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domain
63 d1dxla3
not modelled
5.2
14
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domainFamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domain
64 d1w2za3
not modelled
5.2
31
Fold: Cystatin-likeSuperfamily: Amine oxidase N-terminal regionFamily: Amine oxidase N-terminal region
65 c3qkgA_
not modelled
5.2
11
PDB header: immune systemChain: A: PDB Molecule: protein ambp;PDBTitle: crystal structure of alpha-1-microglobulin at 2.3 a resolution
66 c2r6oB_
not modelled
5.2
9
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative diguanylate cyclase/phosphodiesterase (ggdef & ealPDBTitle: crystal structure of putative diguanylate cyclase/phosphodiesterase2 from thiobacillus denitrificans
67 c4wseA_
not modelled
5.1
30
PDB header: transferaseChain: A: PDB Molecule: putative poly(a) polymerase catalytic subunit;PDBTitle: crystal structure of the mimivirus polyadenylate synthase