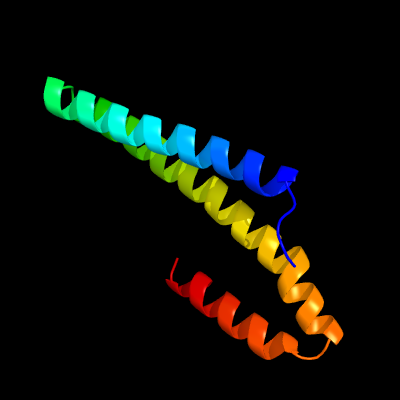
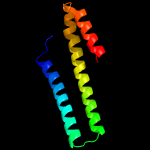
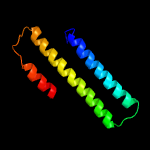
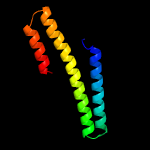
1 c4m1pA_
100.0
28
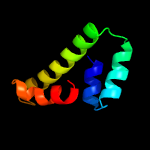
PDB header: transcription repressorChain: A: PDB Molecule: copper-sensitive operon repressor (csor);PDBTitle: crystal structure of the copper-sensing repressor csor with cu(i) from2 geobacillus thermodenitrificans ng80-2
2 c5lbmD_
100.0
22
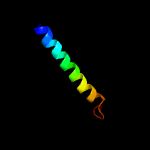
PDB header: transcriptionChain: D: PDB Molecule: transcriptional repressor frmr;PDBTitle: the asymmetric tetrameric structure of the formaldehyde sensing2 transcriptional repressor frmr from escherichia coli
3 c5fmnB_
100.0
35
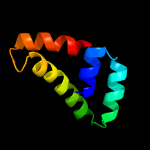
PDB header: dna binding proteinChain: B: PDB Molecule: inrs;PDBTitle: the nickel-responsive transcriptional regulator inrs
4 c5lcyD_
100.0
24
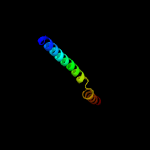
PDB header: transcriptionChain: D: PDB Molecule: frmr;PDBTitle: formaldehyde-responsive regulator frmr e64h variant from salmonella2 enterica serovar typhimurium
5 c4adzA_
99.9
29
PDB header: transcriptionChain: A: PDB Molecule: csor;PDBTitle: crystal structure of the apo form of a copper-sensitive operon2 regulator (csor) protein from streptomyces lividans
6 c2hh7A_
99.9
32
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein csor;PDBTitle: crystal structure of cu(i) bound csor from mycobacterium tuberculosis.
7 c3aaiB_
99.9
34
PDB header: transcriptionChain: B: PDB Molecule: copper homeostasis operon regulatory protein;PDBTitle: x-ray crystal structure of csor from thermus thermophilus hb8
8 c2p5tA_
36.2
38
PDB header: transcription regulatorChain: A: PDB Molecule: putative transcriptional regulator peza;PDBTitle: molecular and structural characterization of the pezat chromosomal2 toxin-antitoxin system of the human pathogen streptococcus pneumoniae
9 c4aj5M_
34.0
15
PDB header: cell cycleChain: M: PDB Molecule: spindle and kinetochore-associated protein 2;PDBTitle: crystal structure of the ska core complex
10 c3zs9D_
31.5
33
PDB header: hydrolase/transport proteinChain: D: PDB Molecule: golgi to er traffic protein 2;PDBTitle: s. cerevisiae get3-adp-alf4- complex with a cytosolic get2 fragment
11 c5frhA_
31.4
12
PDB header: transcriptionChain: A: PDB Molecule: anti-sigma factor rsra;PDBTitle: solution structure of oxidised rsra
12 c1paqA_
29.7
15
PDB header: translationChain: A: PDB Molecule: translation initiation factor eif-2b epsilonPDBTitle: crystal structure of the catalytic fragment of eukaryotic2 initiation factor 2b epsilon
13 d1paqa_
29.7
15
Fold: alpha-alpha superhelixSuperfamily: ARM repeatFamily: MIF4G domain-like
14 d1ivsa1
19.9
6
Fold: Long alpha-hairpinSuperfamily: tRNA-binding armFamily: Valyl-tRNA synthetase (ValRS) C-terminal domain
15 c3juiA_
19.1
15
PDB header: translationChain: A: PDB Molecule: translation initiation factor eif-2b subunit epsilon;PDBTitle: crystal structure of the c-terminal domain of human translation2 initiation factor eif2b epsilon subunit
16 c4u8us_
18.8
4
PDB header: oxygen storage/transportChain: S: PDB Molecule: globin d chain;PDBTitle: the crystallographic structure of the giant hemoglobin from2 glossoscolex paulistus at 3.2 a resolution.
17 c2nv2U_
18.5
18
PDB header: lyase/transferaseChain: U: PDB Molecule: pyridoxal biosynthesis lyase pdxs;PDBTitle: structure of the plp synthase complex pdx1/2 (yaad/e) from bacillus2 subtilis
18 c4fc9B_
18.0
27
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: structure of the c-terminal domain of the type iii effector xcv32202 (xopl)
19 c2qycA_
17.4
18
PDB header: unknown functionChain: A: PDB Molecule: ferredoxin-like protein;PDBTitle: crystal structure of a dimeric ferredoxin-like protein (bb1511) from2 bordetella bronchiseptica rb50 at 1.90 a resolution
20 d1kkga_
14.8
27
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Ribosome-binding factor A, RbfAFamily: Ribosome-binding factor A, RbfA
21 c4n6jA_
not modelled
14.3
21
PDB header: signaling proteinChain: A: PDB Molecule: striatin-3;PDBTitle: crystal structure of human striatin-3 coiled coil domain
22 c4n6jB_
not modelled
13.9
21
PDB header: signaling proteinChain: B: PDB Molecule: striatin-3;PDBTitle: crystal structure of human striatin-3 coiled coil domain
23 c5vf3Z_
not modelled
12.8
31
PDB header: virusChain: Z: PDB Molecule: highly immunogenic outer capsid protein;PDBTitle: bacteriophage t4 isometric capsid
24 c5an5J_
not modelled
12.6
27
PDB header: cell cycleChain: J: PDB Molecule: cell cycle protein gpsb;PDBTitle: b. subtilis gpsb c-terminal domain
25 c2n5xA_
not modelled
12.2
8
PDB header: chaperoneChain: A: PDB Molecule: hsp90 co-chaperone cdc37;PDBTitle: c-terminal domain of cdc37 cochaperone
26 d1josa_
not modelled
11.9
18
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Ribosome-binding factor A, RbfAFamily: Ribosome-binding factor A, RbfA
27 c4lh9A_
not modelled
11.5
14
PDB header: transcriptionChain: A: PDB Molecule: heterocyst differentiation control protein;PDBTitle: crystal structure of the refolded hood domain (asp256-gly295) of hetr
28 c4n5bF_
not modelled
10.3
10
PDB header: viral proteinChain: F: PDB Molecule: phosphoprotein;PDBTitle: crystal structure of the nipah virus phosphoprotein tetramerization2 domain
29 c2gtlO_
not modelled
9.6
10
PDB header: oxygen storage/transportChain: O: PDB Molecule: extracellular hemoglobin linker l3 subunit;PDBTitle: lumbricus erythrocruorin at 3.5a resolution
30 d2igsa1
not modelled
9.2
27
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: PA2222-like
31 c2mhwA_
not modelled
8.4
21
PDB header: antimicrobial protein, membrane proteinChain: A: PDB Molecule: antimicrobial peptide;PDBTitle: the solution nmr structure of maximin-4 in sds micelles
32 c3femB_
not modelled
8.2
9
PDB header: biosynthetic protein, transferaseChain: B: PDB Molecule: pyridoxine biosynthesis protein snz1;PDBTitle: structure of the synthase subunit pdx1.1 (snz1) of plp synthase from2 saccharomyces cerevisiae
33 c6hyeF_
not modelled
8.1
18
PDB header: plant proteinChain: F: PDB Molecule: pyridoxal 5'-phosphate synthase subunit pdx1.3;PDBTitle: pdx1.2/pdx1.3 complex (pdx1.3:k97a)
34 c2ovcA_
not modelled
8.1
10
PDB header: transport proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily kqt member 4;PDBTitle: crystal structure of a coiled-coil tetramerization domain from kv7.42 channels
35 d1q4ra_
not modelled
8.0
5
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Plant stress-induced protein
36 c4p3gC_
not modelled
8.0
12
PDB header: rna binding proteinChain: C: PDB Molecule: signal recognition particle subunit srp68;PDBTitle: structure of the srp68-rbd from chaetomium thermophilum
37 c2i9iA_
not modelled
7.4
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of helicobacter pylori protein hp0492
38 d2i9ia1
not modelled
7.4
11
Fold: Anticodon-binding domain-likeSuperfamily: XCC0632-likeFamily: NLBH-like
39 c2kzfA_
not modelled
6.9
23
PDB header: ribosomal proteinChain: A: PDB Molecule: ribosome-binding factor a;PDBTitle: solution nmr structure of the thermotoga maritima protein tm0855 a2 putative ribosome binding factor a
40 d1tr0a_
not modelled
6.8
9
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Plant stress-induced protein
41 c3cveC_
not modelled
6.8
21
PDB header: signaling proteinChain: C: PDB Molecule: homer protein homolog 1;PDBTitle: crystal structure of the carboxy terminus of homer1
42 c5y02D_
not modelled
6.7
14
PDB header: lyaseChain: D: PDB Molecule: hydroxynitrile lyase;PDBTitle: c-terminal peptide depleted mutant of hydroxynitrile lyase from2 passiflora edulis (pehnl) bound with (r)-mandelonitrile
43 c1cffB_
not modelled
6.6
40
PDB header: calmodulinChain: B: PDB Molecule: calcium pump;PDBTitle: nmr solution structure of a complex of calmodulin with a binding2 peptide of the ca2+-pump
44 c6hxgE_
not modelled
6.3
18
PDB header: plant proteinChain: E: PDB Molecule: pyridoxal 5'-phosphate synthase-like subunit pdx1.2;PDBTitle: pdx1.2/pdx1.3 complex (intermediate)
45 c3ibwA_
not modelled
6.1
26
PDB header: transferaseChain: A: PDB Molecule: gtp pyrophosphokinase;PDBTitle: crystal structure of the act domain from gtp pyrophosphokinase of2 chlorobium tepidum. northeast structural genomics consortium target3 ctr148a
46 c4yioB_
not modelled
6.1
13
PDB header: oxidoreductaseChain: B: PDB Molecule: superoxide dismutase;PDBTitle: x-ray structure of the iron/manganese cambialistic superoxide2 dismutase from streptococcus thermophilus
47 c3vdoB_
not modelled
6.1
6
PDB header: dna binding protein/protein bindingChain: B: PDB Molecule: anti-sigma-k factor rska;PDBTitle: structure of extra-cytoplasmic function(ecf) sigma factor sigk in2 complex with its negative regulator rska from mycobacterium3 tuberculosis
48 d1tiza_
not modelled
6.0
9
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like
49 c5b0aA_
not modelled
5.9
0
PDB header: lyaseChain: A: PDB Molecule: olivetolic acid cyclase;PDBTitle: polyketide cyclase oac from cannabis sativa, h5q mutant
50 c3rkgA_
not modelled
5.7
4
PDB header: metal transportChain: A: PDB Molecule: magnesium transporter mrs2, mitochondrial;PDBTitle: structural and functional characterization of the yeast mg2+ channel2 mrs2
51 c6bp6A_
not modelled
5.5
30
PDB header: endocytosisChain: A: PDB Molecule: comm domain-containing protein 9;PDBTitle: crystal structure of commd9 comm domain
52 c3nepX_
not modelled
5.5
17
PDB header: oxidoreductaseChain: X: PDB Molecule: malate dehydrogenase;PDBTitle: 1.55a resolution structure of malate dehydrogenase from salinibacter2 ruber
53 d1pa4a_
not modelled
5.3
17
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Ribosome-binding factor A, RbfAFamily: Ribosome-binding factor A, RbfA
54 d1gtda_
not modelled
5.3
17
Fold: PurS-likeSuperfamily: PurS-likeFamily: PurS subunit of FGAM synthetase
55 d1j2jb_
not modelled
5.3
16
Fold: Spectrin repeat-likeSuperfamily: GAT-like domainFamily: GAT domain
56 c2xa0C_
not modelled
5.3
26
PDB header: apoptosisChain: C: PDB Molecule: apoptosis regulator bax;PDBTitle: crystal structure of bcl-2 in complex with a bax bh32 peptide
57 c4oe8C_
not modelled
5.1
19
PDB header: immune systemChain: C: PDB Molecule: alphabody ma12;PDBTitle: interleukin-23 complex with an antagonistic alphabody, crystal form 1