1 c4kxrC_
100.0
99
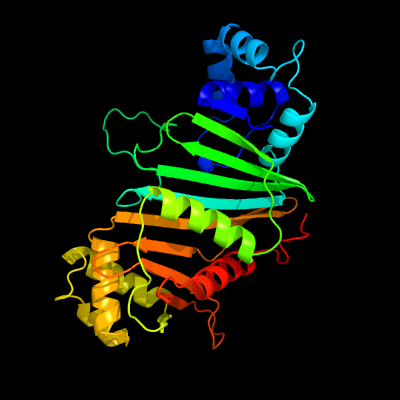
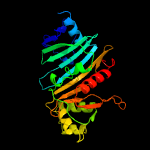
PDB header: protein transportChain: C: PDB Molecule: espg5;PDBTitle: structure of the mycobacterium tuberculosis type vii secretion system2 chaperone espg5 in complex with pe25-ppe41 dimer
2 c4l4wB_
100.0
21
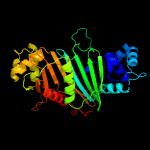
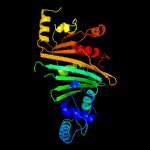
PDB header: protein transportChain: B: PDB Molecule: espg3;PDBTitle: structure of espg3 chaperone from the type vii (esx-3) secretion2 system
3 c4w4iA_
100.0
21
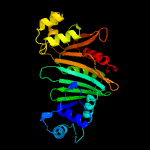
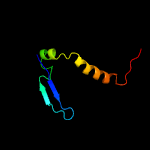
PDB header: protein transportChain: A: PDB Molecule: esx-3 secretion-associated protein espg3;PDBTitle: crystal structure of espg3 from the esx-3 type vii secretion system of2 m. tuberculosis
4 c4rclB_
100.0
20
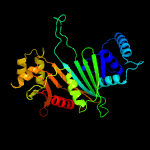
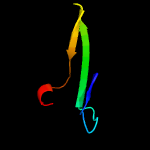
PDB header: chaperoneChain: B: PDB Molecule: espg3;PDBTitle: structure of espg3 chaperone from the type vii (esx-3) secretion2 system, space group p43212
5 c5vbaA_
100.0
26
PDB header: chaperone, hydrolaseChain: A: PDB Molecule: lysozyme, esx-1 secretion-associated protein espg1 chimera;PDBTitle: structure of espg1 chaperone from the type vii (esx-1) secretion2 system determined with the assistance of n-terminal t4 lysozyme3 fusion
6 d1w79a1
24.6
16
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: Dac-like
7 c4rdkB_
17.6
21
PDB header: viral proteinChain: B: PDB Molecule: capsid;PDBTitle: crystal structure of norovirus boxer p domain in complex with lewis b2 tetrasaccharide
8 c3a3eB_
15.5
20
PDB header: hydrolaseChain: B: PDB Molecule: penicillin-binding protein 4;PDBTitle: crystal structure of penicillin binding protein 4 (dacb) from2 haemophilus influenzae, complexed with novel beta-lactam (cmv)
9 d1hynp_
13.8
15
Fold: Phoshotransferase/anion transport proteinSuperfamily: Phoshotransferase/anion transport proteinFamily: Anion transport protein, cytoplasmic domain
10 d1ppva_
13.1
12
Fold: NudixSuperfamily: NudixFamily: IPP isomerase-like
11 c1hynQ_
11.8
15
PDB header: membrane proteinChain: Q: PDB Molecule: band 3 anion transport protein;PDBTitle: crystal structure of the cytoplasmic domain of human erythrocyte band-2 3 protein
12 d1rk8c_
11.5
45
Fold: WW domain-likeSuperfamily: Pym (Within the bgcn gene intron protein, WIBG), N-terminal domainFamily: Pym (Within the bgcn gene intron protein, WIBG), N-terminal domain
13 c1rk8C_
11.5
45
PDB header: translationChain: C: PDB Molecule: within the bgcn gene intron protein;PDBTitle: structure of the cytosolic protein pym bound to the mago-2 y14 core of the exon junction complex
14 c2ei5B_
11.2
23
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein ttha0061;PDBTitle: crystal structure of hypothetical protein(ttha0061) from thermus2 thermophilus
15 d1u94a2
10.3
25
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain
16 d1mo6a2
10.2
38
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain
17 d2fvva1
10.1
21
Fold: NudixSuperfamily: NudixFamily: MutT-like
18 c2fvvA_
10.1
21
PDB header: hydrolaseChain: A: PDB Molecule: diphosphoinositol polyphosphate phosphohydrolase 1;PDBTitle: human diphosphoinositol polyphosphate phosphohydrolase 1
19 c2v4oB_
10.0
25
PDB header: hydrolaseChain: B: PDB Molecule: multifunctional protein sur e;PDBTitle: crystal structure of salmonella typhimurium sure at 2.752 angstrom resolution in monoclinic form
20 d1rrqa2
9.7
11
Fold: NudixSuperfamily: NudixFamily: MutY C-terminal domain-like
21 d1kdga2
not modelled
9.4
21
Fold: FAD-linked reductases, C-terminal domainSuperfamily: FAD-linked reductases, C-terminal domainFamily: GMC oxidoreductases
22 d1xp8a2
not modelled
9.2
13
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain
23 d1ubea2
not modelled
9.1
38
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain
24 c2ly2A_
not modelled
8.7
26
PDB header: rna binding proteinChain: A: PDB Molecule: tudor domain-containing protein 7;PDBTitle: nmr structure of the second and third lotus domains of tudor domain-2 containing protein 7 (nmr ensemble overlay for lotus #3)
25 d2ex2a1
not modelled
8.6
17
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: Dac-like
26 d1w5da1
not modelled
8.3
9
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: Dac-like
27 d1v7ba2
not modelled
8.1
15
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain
28 c4dibF_
not modelled
7.5
33
PDB header: oxidoreductaseChain: F: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: the crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 bacillus anthracis str. sterne
29 c2x5kO_
not modelled
7.4
33
PDB header: oxidoreductaseChain: O: PDB Molecule: d-erythrose-4-phosphate dehydrogenase;PDBTitle: structure of an active site mutant of the d-erythrose-4-phosphate2 dehydrogenase from e. coli
30 c5fgoA_
not modelled
7.3
7
PDB header: dna binding proteinChain: A: PDB Molecule: cg1507-pb, isoform b;PDBTitle: crystal structure of d. melanogaster pur-alpha repeat iii.
31 c2eh6A_
not modelled
7.2
10
PDB header: transferaseChain: A: PDB Molecule: acetylornithine aminotransferase;PDBTitle: crystal structure of acetylornithine aminotransferase from aquifex2 aeolicus vf5
32 c2o1cB_
not modelled
7.1
15
PDB header: hydrolaseChain: B: PDB Molecule: datp pyrophosphohydrolase;PDBTitle: structure of the e. coli dihydroneopterin triphosphate2 pyrophosphohydrolase
33 c4zyeA_
not modelled
7.1
21
PDB header: transferaseChain: A: PDB Molecule: methylated-dna--protein-cysteine methyltransferase;PDBTitle: crystal structure of sulfolobus solfataricus o6-methylguanine2 methyltransferase
34 c2i5pO_
not modelled
7.0
22
PDB header: oxidoreductaseChain: O: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase 1;PDBTitle: crystal structure of glyceraldehyde-3-phosphate2 dehydrogenase isoform 1 from k. marxianus
35 c5jyfB_
not modelled
6.9
22
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: structures of streptococcus agalactiae gbs gapdh in different2 enzymatic states
36 c4qx6A_
not modelled
6.7
22
PDB header: oxidoreductaseChain: A: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 streptococcus agalactiae nem316 at 2.46 angstrom resolution
37 c1obfO_
not modelled
6.5
33
PDB header: glycolytic pathwayChain: O: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: the crystal structure of glyceraldehyde 3-phosphate2 dehydrogenase from alcaligenes xylosoxidans at 1.7 a3 resolution.
38 c5ur0B_
not modelled
6.4
22
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystallographic structure of glyceraldehyde-3-phosphate dehydrogenase2 from naegleria gruberi
39 c3h9eO_
not modelled
6.4
33
PDB header: oxidoreductaseChain: O: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase, testis-specific;PDBTitle: crystal structure of human sperm-specific glyceraldehyde-3-phosphate2 dehydrogenase (gapds) complex with nad and phosphate
40 c5j9gB_
not modelled
6.4
11
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-p dehydrogenase;PDBTitle: structure of lactobacillus acidophilus glyceraldehyde-3-phosphate2 dehydrogenase at 2.21 angstrom resolution
41 c1s7cA_
not modelled
6.4
33
PDB header: structural genomics, oxidoreductaseChain: A: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase a;PDBTitle: crystal structure of mes buffer bound form of glyceraldehyde 3-2 phosphate dehydrogenase from escherichia coli
42 c2b4rQ_
not modelled
6.3
33
PDB header: oxidoreductaseChain: Q: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 plasmodium falciparum at 2.25 angstrom resolution reveals intriguing3 extra electron density in the active site
43 c3hq4R_
not modelled
6.3
11
PDB header: oxidoreductaseChain: R: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase 1;PDBTitle: crystal structure of c151s mutant of glyceraldehyde-3-phosphate2 dehydrogenase 1 (gapdh1) complexed with nad from staphylococcus3 aureus mrsa252 at 2.2 angstrom resolution
44 d1v8ya_
not modelled
6.3
13
Fold: NudixSuperfamily: NudixFamily: MutT-like
45 d1g26a_
not modelled
6.2
50
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Granulin repeatFamily: Granulin repeat
46 c2d2iO_
not modelled
6.2
22
PDB header: oxidoreductaseChain: O: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: crystal structure of nadp-dependent glyceraldehyde-3-2 phosphate dehydrogenase from synechococcus sp. complexed3 with nadp+
47 c3cieC_
not modelled
6.2
33
PDB header: oxidoreductaseChain: C: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde 3-phosphate2 dehydrogenase from cryptosporidium parvum
48 c5ld5C_
not modelled
6.2
22
PDB header: oxidoreductaseChain: C: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of a bacterial dehydrogenase at 2.19 angstroms2 resolution
49 c3nx3A_
not modelled
6.1
12
PDB header: transferaseChain: A: PDB Molecule: acetylornithine aminotransferase;PDBTitle: crystal structure of acetylornithine aminotransferase (argd) from2 campylobacter jejuni
50 c3b20R_
not modelled
6.1
22
PDB header: oxidoreductaseChain: R: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase (nadp+);PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase2 complexed with nadfrom synechococcus elongatus"
51 c2ep7B_
not modelled
6.0
22
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: structural study of project id aq_1065 from aquifex aeolicus vf5
52 c1ihxD_
not modelled
6.0
33
PDB header: oxidoreductaseChain: D: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: crystal structure of two d-glyceraldehyde-3-phosphate2 dehydrogenase complexes: a case of asymmetry
53 c1hdgO_
not modelled
6.0
33
PDB header: oxidoreductase (aldehy(d)-nad(a))Chain: O: PDB Molecule: holo-d-glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: the crystal structure of holo-glyceraldehyde-3-phosphate dehydrogenase2 from the hyperthermophilic bacterium thermotoga maritima at 2.53 angstroms resolution
54 d1qbea_
not modelled
6.0
20
Fold: RNA bacteriophage capsid proteinSuperfamily: RNA bacteriophage capsid proteinFamily: RNA bacteriophage capsid protein
55 c4o9lA_
not modelled
5.9
17
PDB header: antiviral proteinChain: A: PDB Molecule: mitochondrial antiviral signaling protein (mavs);PDBTitle: crystal structure of horse mavs card domain mutant e26r
56 c3docD_
not modelled
5.9
44
PDB header: oxidoreductaseChain: D: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: crystal structure of trka glyceraldehyde-3-phosphate dehydrogenase2 from brucella melitensis
57 c3hjaB_
not modelled
5.9
44
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 borrelia burgdorferi
58 c2pkrI_
not modelled
5.8
22
PDB header: oxidoreductaseChain: I: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase aor;PDBTitle: crystal structure of (a+cte)4 chimeric form of photosyntetic2 glyceraldehyde-3-phosphate dehydrogenase, complexed with nadp
59 c1cerC_
not modelled
5.8
22
PDB header: oxidoreductase (aldehyde(d)-nad(a))Chain: C: PDB Molecule: holo-d-glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: determinants of enzyme thermostability observed in the2 molecular structure of thermus aquaticus d-glyceraldehyde-3 3-phosphate dehydrogenase at 2.5 angstroms resolution
60 c6ok4A_
not modelled
5.8
22
PDB header: oxidoreductaseChain: A: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase (gapdh)2 from chlamydia trachomatis with bound nad
61 c1rm4O_
not modelled
5.7
22
PDB header: oxidoreductaseChain: O: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase a;PDBTitle: crystal structure of recombinant photosynthetic glyceraldehyde-3-2 phosphate dehydrogenase a4 isoform, complexed with nadp
62 d1mgta1
not modelled
5.7
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domainFamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domain
63 c3sthA_
not modelled
5.7
33
PDB header: oxidoreductaseChain: A: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 toxoplasma gondii
64 d1vhza_
not modelled
5.6
2
Fold: NudixSuperfamily: NudixFamily: MutT-like
65 c1i32D_
not modelled
5.6
22
PDB header: oxidoreductaseChain: D: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: leishmania mexicana glyceraldehyde-3-phosphate2 dehydrogenase in complex with inhibitors
66 c3u5gK_
not modelled
5.4
21
PDB header: ribosomeChain: K: PDB Molecule: 40s ribosomal protein s10-a;PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution. this2 entry contains proteins of the 40s subunit, ribosome b
67 c3i4jC_
not modelled
5.2
16
PDB header: transferaseChain: C: PDB Molecule: aminotransferase, class iii;PDBTitle: crystal structure of aminotransferase, class iii from deinococcus2 radiodurans