| 1 |
|
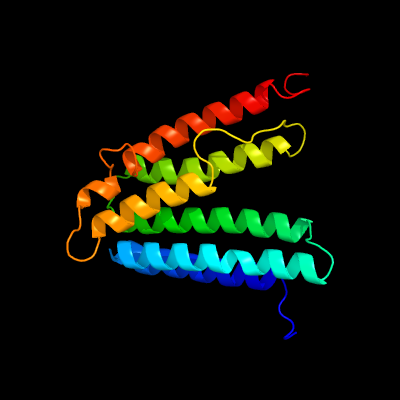
PDB 6h59 chain B
Region: 9 - 192
Aligned: 178
Modelled: 184
Confidence: 99.9%
Identity: 15%
PDB header:transferase
Chain: B: PDB Molecule:cdp-diacylglycerol--inositol 3-phosphatidyltransferase;
PDBTitle: crystal structure of mycobacterium tuberculosis phosphatidylinositol2 phosphate synthase (pgsa1) with cdp-dag bound
Phyre2
| 2 |
|
PDB 5d92 chain B
Region: 14 - 187
Aligned: 169
Modelled: 174
Confidence: 99.9%
Identity: 20%
PDB header:membrane protein
Chain: B: PDB Molecule:af2299 protein,phosphatidylinositol synthase;
PDBTitle: structure of a phosphatidylinositolphosphate (pip) synthase from2 renibacterium salmoninarum
Phyre2
| 3 |
|
PDB 4mnd chain A
Region: 14 - 177
Aligned: 162
Modelled: 164
Confidence: 99.9%
Identity: 15%
PDB header:transferase
Chain: A: PDB Molecule:ctp l-myo-inositol-1-phosphate cytidylyltransferase/cdp-l-
PDBTitle: crystal structure of archaeoglobus fulgidus ipct-dipps bifunctional2 membrane protein
Phyre2
| 4 |
|
PDB 4o6m chain A
Region: 14 - 186
Aligned: 159
Modelled: 159
Confidence: 99.9%
Identity: 15%
PDB header:transferase
Chain: A: PDB Molecule:af2299, a cdp-alcohol phosphotransferase;
PDBTitle: structure of af2299, a cdp-alcohol phosphotransferase (cmp-bound)
Phyre2
| 5 |
|
PDB 4k1c chain B
Region: 22 - 209
Aligned: 186
Modelled: 188
Confidence: 48.8%
Identity: 11%
PDB header:membrane protein/metal transport
Chain: B: PDB Molecule:vacuolar calcium ion transporter;
PDBTitle: vcx1 calcium/proton exchanger
Phyre2
| 6 |
|
PDB 4k1c chain A
Region: 22 - 208
Aligned: 185
Modelled: 187
Confidence: 38.0%
Identity: 11%
PDB header:membrane protein/metal transport
Chain: A: PDB Molecule:vacuolar calcium ion transporter;
PDBTitle: vcx1 calcium/proton exchanger
Phyre2
| 7 |
|
PDB 6mjb chain C
Region: 60 - 77
Aligned: 18
Modelled: 18
Confidence: 28.6%
Identity: 33%
PDB header:cell cycle
Chain: C: PDB Molecule:kinetochore-associated protein dsn1;
PDBTitle: structure of candida glabrata csm1:dsn1(14-72) complex
Phyre2
| 8 |
|
PDB 5guf chain A
Region: 18 - 116
Aligned: 95
Modelled: 99
Confidence: 17.4%
Identity: 14%
PDB header:transferase
Chain: A: PDB Molecule:cdp-archaeol synthase;
PDBTitle: structural insight into an intramembrane enzyme for archaeal membrane2 lipids biosynthesis
Phyre2
| 9 |
|
PDB 2rkh chain A
Region: 46 - 79
Aligned: 34
Modelled: 34
Confidence: 11.0%
Identity: 29%
PDB header:transcription
Chain: A: PDB Molecule:putative apha-like transcription factor;
PDBTitle: crystal structure of a putative apha-like transcription factor2 (zp_00208345.1) from magnetospirillum magnetotacticum ms-1 at 2.00 a3 resolution
Phyre2
| 10 |
|
PDB 3eh4 chain A
Region: 162 - 209
Aligned: 48
Modelled: 48
Confidence: 10.8%
Identity: 17%
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome c oxidase subunit 1;
PDBTitle: structure of the reduced form of cytochrome ba3 oxidase from thermus2 thermophilus
Phyre2
| 11 |
|
PDB 2voy chain G
Region: 63 - 90
Aligned: 28
Modelled: 28
Confidence: 10.8%
Identity: 21%
PDB header:hydrolase
Chain: G: PDB Molecule:sarcoplasmic/endoplasmic reticulum calcium atpase 1;
PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
Phyre2
| 12 |
|
PDB 1xme chain A domain 1
Region: 162 - 209
Aligned: 48
Modelled: 48
Confidence: 9.5%
Identity: 17%
Fold: Cytochrome c oxidase subunit I-like
Superfamily: Cytochrome c oxidase subunit I-like
Family: Cytochrome c oxidase subunit I-like
Phyre2
| 13 |
|
PDB 2le7 chain A
Region: 57 - 70
Aligned: 14
Modelled: 14
Confidence: 8.2%
Identity: 43%
PDB header:transport protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily h member 2;
PDBTitle: solution nmr structure of the s4s5 linker of herg potassium channel
Phyre2
| 14 |
|
PDB 2n28 chain A
Region: 163 - 209
Aligned: 47
Modelled: 47
Confidence: 7.2%
Identity: 15%
PDB header:viral protein
Chain: A: PDB Molecule:protein vpu;
PDBTitle: solid-state nmr structure of vpu
Phyre2
| 15 |
|
PDB 4arp chain B
Region: 46 - 75
Aligned: 30
Modelled: 30
Confidence: 5.6%
Identity: 13%
PDB header:hydrolase
Chain: B: PDB Molecule:pesticin;
PDBTitle: structure of the inactive pesticin e178a mutant
Phyre2