| 1 |
|
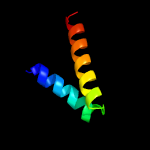
PDB 3cxn chain B
Region: 1 - 190
Aligned: 190
Modelled: 190
Confidence: 100.0%
Identity: 16%
PDB header:chaperone
Chain: B: PDB Molecule:urease accessory protein uref;
PDBTitle: structure of the urease accessory protein uref from helicobacter2 pylori
Phyre2
| 2 |
|
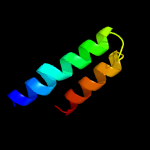
PDB 3w3f chain B
Region: 88 - 156
Aligned: 69
Modelled: 69
Confidence: 16.2%
Identity: 17%
PDB header:biosynthetic protein
Chain: B: PDB Molecule:uncharacterized protein blr2150;
PDBTitle: crystal structure of ent-kaurene synthase bjks from bradyrhizobium2 japonicum
Phyre2
| 3 |
|
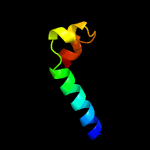
PDB 2mtx chain A
Region: 12 - 20
Aligned: 9
Modelled: 9
Confidence: 15.0%
Identity: 56%
PDB header:protein binding
Chain: A: PDB Molecule:apical membrane antigen-1;
PDBTitle: protection against experimental p. falciparum malaria is associated2 with short ama-1 peptide analogue alpha-helical structures
Phyre2
| 4 |
|
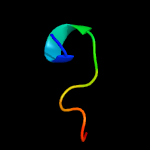
PDB 3mx7 chain A
Region: 16 - 27
Aligned: 12
Modelled: 12
Confidence: 13.5%
Identity: 8%
PDB header:apoptosis
Chain: A: PDB Molecule:fas apoptotic inhibitory molecule 1;
PDBTitle: crystal structure analysis of human faim-ntd
Phyre2
| 5 |
|
PDB 5mk5 chain A
Region: 161 - 200
Aligned: 40
Modelled: 40
Confidence: 11.8%
Identity: 15%
PDB header:hydrolase
Chain: A: PDB Molecule:bloom syndrome protein;
PDBTitle: structures of dhbn domain of human blm helicase
Phyre2
| 6 |
|
PDB 6iuh chain C
Region: 158 - 182
Aligned: 25
Modelled: 25
Confidence: 10.9%
Identity: 16%
PDB header:protein binding
Chain: C: PDB Molecule:liprin-alpha-2;
PDBTitle: crystal structure of git1 pbd domain in complex with liprin-alpha2
Phyre2
| 7 |
|
PDB 2odm chain A
Region: 135 - 175
Aligned: 40
Modelled: 41
Confidence: 10.3%
Identity: 13%
PDB header:unknown function
Chain: A: PDB Molecule:upf0358 protein mw0995;
PDBTitle: crystal structure of s. aureus ylan, an essential leucine rich protein2 involved in the control of cell shape
Phyre2
| 8 |
|
PDB 1yqg chain A domain 1
Region: 140 - 181
Aligned: 41
Modelled: 42
Confidence: 10.0%
Identity: 20%
Fold: 6-phosphogluconate dehydrogenase C-terminal domain-like
Superfamily: 6-phosphogluconate dehydrogenase C-terminal domain-like
Family: ProC C-terminal domain-like
Phyre2
| 9 |
|
PDB 2gbo chain A domain 1
Region: 135 - 175
Aligned: 40
Modelled: 41
Confidence: 9.5%
Identity: 20%
Fold: Open three-helical up-and-down bundle
Superfamily: EF2458-like
Family: EF2458-like
Phyre2
| 10 |
|
PDB 2gbo chain B
Region: 135 - 175
Aligned: 40
Modelled: 41
Confidence: 9.5%
Identity: 20%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:upf0358 protein ef2458;
PDBTitle: protein of unknown function ef2458 from enterococcus faecalis
Phyre2
| 11 |
|
PDB 2f4m chain A domain 1
Region: 112 - 133
Aligned: 22
Modelled: 22
Confidence: 9.4%
Identity: 14%
Fold: Cysteine proteinases
Superfamily: Cysteine proteinases
Family: Transglutaminase core
Phyre2
| 12 |
|
PDB 3f5h chain B
Region: 73 - 94
Aligned: 22
Modelled: 22
Confidence: 9.0%
Identity: 18%
PDB header:protein binding
Chain: B: PDB Molecule:type i polyketide synthase pikaiii, type i polyketide
PDBTitle: crystal structure of fused docking domains from pikaiii and pikaiv of2 the pikromycin polyketide synthase
Phyre2
| 13 |
|
PDB 5lut chain G
Region: 165 - 200
Aligned: 36
Modelled: 36
Confidence: 8.7%
Identity: 11%
PDB header:transferase
Chain: G: PDB Molecule:blm helicase;
PDBTitle: structures of dhbn domain of gallus gallus blm helicase
Phyre2
| 14 |
|
PDB 3suj chain A
Region: 13 - 19
Aligned: 7
Modelled: 7
Confidence: 7.7%
Identity: 29%
PDB header:unknown function
Chain: A: PDB Molecule:cerato-platanin 1;
PDBTitle: crystal structure of cerato-platanin 1 from m. perniciosa (mpcp1)
Phyre2
| 15 |
|
PDB 4zzp chain B
Region: 8 - 19
Aligned: 12
Modelled: 12
Confidence: 7.2%
Identity: 33%
PDB header:hydrolase
Chain: B: PDB Molecule:cellulose 1,4-beta-cellobiosidase;
PDBTitle: dictyostelium purpureum cellobiohydrolase cel7a apo structure
Phyre2
| 16 |
|
PDB 3shp chain A
Region: 10 - 19
Aligned: 10
Modelled: 10
Confidence: 7.0%
Identity: 50%
PDB header:transferase
Chain: A: PDB Molecule:putative acetyltransferase sthe_0691;
PDBTitle: crystal structure of putative acetyltransferase from sphaerobacter2 thermophilus dsm 20745
Phyre2
| 17 |
|
PDB 1iw4 chain A
Region: 18 - 25
Aligned: 8
Modelled: 8
Confidence: 6.9%
Identity: 38%
Fold: Kazal-type serine protease inhibitors
Superfamily: Kazal-type serine protease inhibitors
Family: Ovomucoid domain III-like
Phyre2
| 18 |
|
PDB 4yf1 chain D
Region: 118 - 148
Aligned: 31
Modelled: 31
Confidence: 5.7%
Identity: 13%
PDB header:hydrolase
Chain: D: PDB Molecule:lmo0812 protein;
PDBTitle: 1.85 angstrom crystal structure of lmo0812 from listeria monocytogenes2 egd-e
Phyre2