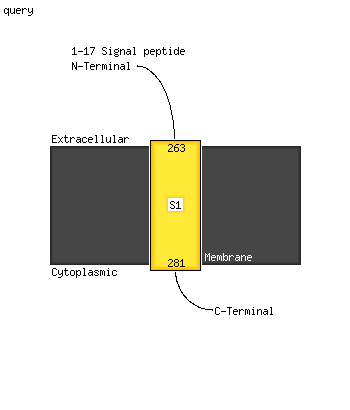
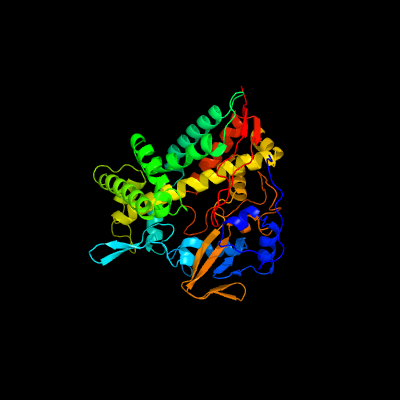
| 1 | c4l0eA_
|
|
 |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:p450 monooxygenase;
PDBTitle: structure of p450sky (cyp163b3), a cytochrome p450 from skyllamycin2 biosynthesis (heme-coordinated expression tag)
|
|
|
|
| 2 | c2wivA_
|
|
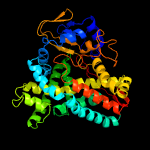 |
100.0 |
24 |
PDB header:electron transport
Chain: A: PDB Molecule:cytochrome p450-like protein xpla;
PDBTitle: cytochrome-p450 xpla heme domain p21
|
|
|
|
| 3 | c3p3oA_
|
|
 |
100.0 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450;
PDBTitle: crystal structure of the cytochrome p450 monooxygenase aurh (ntermii)2 from streptomyces thioluteus
|
|
|
|
| 4 | d1cpta_
|
|
 |
100.0 |
26 |
Fold:Cytochrome P450
Superfamily:Cytochrome P450
Family:Cytochrome P450 |
|
|
|
| 5 | c3bujA_
|
|
 |
100.0 |
28 |
PDB header:metal binding protein
Chain: A: PDB Molecule:calo2;
PDBTitle: crystal structure of calo2
|
|
|
|
| 6 | c5fyfA_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450;
PDBTitle: structure of cyp153a from marinobacter aquaeolei
|
|
|
|
| 7 | c3tktA_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450;
PDBTitle: crystal structure of cyp108d1 from novosphingobium aromaticivorans2 dsm12444
|
|
|
|
| 8 | c5hdiA_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 144;
PDBTitle: structural characterization of cyp144a1, a mycobacterium tuberculosis2 cytochrome p450
|
|
|
|
| 9 | c6giiA_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450;
PDBTitle: the crystal structure of tepidiphilus thermophilus p450 heme domain
|
|
|
|
| 10 | c2fr7A_
|
|
 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative cytochrome p450;
PDBTitle: crystal structure of cytochrome p450 cyp199a2
|
|
|
|
| 11 | c6hqdB_
|
|
 |
100.0 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cytochrome p450;
PDBTitle: cytochrome p450-153 from pseudomonas sp. 19-rlim
|
|
|
|
| 12 | d1q5da_
|
|
 |
100.0 |
29 |
Fold:Cytochrome P450
Superfamily:Cytochrome P450
Family:Cytochrome P450 |
|
|
|
| 13 | c4l36B_
|
|
 |
100.0 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative p450-like protein;
PDBTitle: crystal structure of the cytochrome p450 enzyme txte
|
|
|
|
| 14 | c5li8A_
|
|
 |
100.0 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative cytochrome p450 126;
PDBTitle: crystal structure of mycobacterium tuberculosis cyp126a1 in complex2 with ketoconazole
|
|
|
|
| 15 | c5gweB_
|
|
 |
100.0 |
25 |
PDB header:electron transport
Chain: B: PDB Molecule:cytochrome p450;
PDBTitle: cytochrome p450 crej
|
|
|
|
| 16 | c2xkrA_
|
|
 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative cytochrome p450 142;
PDBTitle: crystal structure of mycobacterium tuberculosis cyp142: a novel2 cholesterol oxidase
|
|
|
|
| 17 | c6bldA_
|
|
 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 268a2 cyp268a2;
PDBTitle: mycobacterium marinum cytochrome p450 cyp268a2 in complex with2 pseudoionone
|
|
|
|
| 18 | c3mgxB_
|
|
 |
100.0 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative p450 monooxygenase;
PDBTitle: crystal structure of p450 oxyd that is involved in the biosynthesis of2 vancomycin-type antibiotics
|
|
|
|
| 19 | c6gk5A_
|
|
 |
100.0 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 cyp267b1 protein;
PDBTitle: crystal structure of cytochrome p450 cyp267b1 from sorangium2 cellulosum so ce56
|
|
|
|
| 20 | c6hqwA_
|
|
 |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450;
PDBTitle: cytochrome p450-153 from novosphingobium aromaticivorans
|
|
|
|
| 21 | c6g71A_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450;
PDBTitle: structure of cyp1232a24 from arthrobacter sp.
|
|
|
| 22 | c5foiB_ |
|
not modelled |
100.0 |
31 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:mycinamicin viii c21 methyl hydroxylase;
PDBTitle: crystal structure of mycinamicin viii c21 methyl hydroxylase mycci2 from micromonospora griseorubida bound to mycinamicin viii
|
|
|
| 23 | c3lxiB_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cytochrome p450;
PDBTitle: crystal structure of camphor-bound cyp101d1
|
|
|
| 24 | c2wm5A_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative cytochrome p450 124;
PDBTitle: x-ray structure of the substrate-free mycobacterium tuberculosis2 cytochrome p450 cyp124
|
|
|
| 25 | c2uvnB_ |
|
not modelled |
100.0 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cytochrome p450 130;
PDBTitle: crystal structure of econazole-bound cyp130 from mycobacterium2 tuberculosis
|
|
|
| 26 | c6dcdA_ |
|
not modelled |
100.0 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 150a6 cyp150a6;
PDBTitle: mycobacterium marinum cytochrome p450 cyp150a6 in the substrate-free2 form
|
|
|
| 27 | c3wvsA_ |
|
not modelled |
100.0 |
31 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative monooxygenase;
PDBTitle: crystal structure of cytochrome p450revi
|
|
|
| 28 | d1re9a_ |
|
not modelled |
100.0 |
24 |
Fold:Cytochrome P450
Superfamily:Cytochrome P450
Family:Cytochrome P450 |
|
|
| 29 | c3rwlA_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 alkane hydroxylase 1 cyp153a7;
PDBTitle: structure of p450pyr hydroxylase
|
|
|
| 30 | c3r9cA_ |
|
not modelled |
100.0 |
34 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 164a2;
PDBTitle: crystal structure of mycobacterium smegmatis cyp164a2 with econazole2 bound
|
|
|
| 31 | c5h1zA_ |
|
not modelled |
100.0 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative cyp alkane hydroxylase cyp153d17;
PDBTitle: cyp153d17 from sphingomonas sp. pamc 26605
|
|
|
| 32 | c2z36A_ |
|
not modelled |
100.0 |
31 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 type compactin 3'',4''-
PDBTitle: crystal structure of cytochrome p450 moxa from nonomuraea2 recticatena (cyp105)
|
|
|
| 33 | c4mm0B_ |
|
not modelled |
100.0 |
31 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:p450-like monooxygenase;
PDBTitle: crystal structure analysis of the putative thioether synthase sgvp2 involved in the tailoring step of griseoviridin
|
|
|
| 34 | c5livC_ |
|
not modelled |
100.0 |
25 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:cytochrome p450 cyp260a1,cytochrome p450 cyp260a1;
PDBTitle: crystal structure of myxobacterial cyp260a1
|
|
|
| 35 | c2xbkA_ |
|
not modelled |
100.0 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pimd protein;
PDBTitle: x-ray structure of the substrate-bound cytochrome p450 pimd - a2 polyene macrolide antibiotic pimaricin epoxidase
|
|
|
| 36 | c3a4hA_ |
|
not modelled |
100.0 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:vitamin d hydroxylase;
PDBTitle: structure of cytochrome p450 vdh from pseudonocardia autotrophica2 (orthorhombic crystal form)
|
|
|
| 37 | c3e5kA_ |
|
not modelled |
100.0 |
35 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 (cytochrome p450 hydroxylase);
PDBTitle: crystal structure of cyp105p1 wild-type 4-phenylimidazole complex
|
|
|
| 38 | c5ncbA_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450;
PDBTitle: crystal structure of amycolatopsis cytochrome p450 gcoa in complex2 with guaiacol.
|
|
|
| 39 | c2zbxA_ |
|
not modelled |
100.0 |
31 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450-su1;
PDBTitle: crystal structure of vitamin d hydroxylase cytochrome p4502 105a1 (wild type) with imidazole bound
|
|
|
| 40 | c3abbA_ |
|
not modelled |
100.0 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 hydroxylase;
PDBTitle: crystal structure of cyp105d6
|
|
|
| 41 | c6hqgB_ |
|
not modelled |
100.0 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cytochrome p450;
PDBTitle: cytochrome p450-153 from phenylobacterium zucineum
|
|
|
| 42 | c5hh3C_ |
|
not modelled |
100.0 |
28 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:oxya protein;
PDBTitle: oxya from actinoplanes teichomyceticus
|
|
|
| 43 | d1lfka_ |
|
not modelled |
100.0 |
29 |
Fold:Cytochrome P450
Superfamily:Cytochrome P450
Family:Cytochrome P450 |
|
|
| 44 | c2z3tD_ |
|
not modelled |
100.0 |
32 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:cytochrome p450;
PDBTitle: crystal structure of substrate free cytochrome p450 stap2 (cyp245a1)
|
|
|
| 45 | d1gwia_ |
|
not modelled |
100.0 |
28 |
Fold:Cytochrome P450
Superfamily:Cytochrome P450
Family:Cytochrome P450 |
|
|
| 46 | c3ejdD_ |
|
not modelled |
100.0 |
33 |
PDB header:oxidoreductase/lipid transport
Chain: D: PDB Molecule:biotin biosynthesis cytochrome p450-like enzyme;
PDBTitle: crystal structure of p450bioi in complex with hexadec-9z-enoic acid2 ligated acyl carrier protein
|
|
|
| 47 | d1z8oa1 |
|
not modelled |
100.0 |
35 |
Fold:Cytochrome P450
Superfamily:Cytochrome P450
Family:Cytochrome P450 |
|
|
| 48 | c4z5pB_ |
|
not modelled |
100.0 |
32 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cytochrome p450 hydroxylase;
PDBTitle: crystal structure of the lnma cytochrome p450 hydroxylase from the2 leinamycin biosynthetic pathway of streptomyces atroolivaceus s-1403 at 1.9 a resolution
|
|
|
| 49 | c3nc7A_ |
|
not modelled |
100.0 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 cypx;
PDBTitle: cyp134a1 2-phenylimidazole bound structure
|
|
|
| 50 | d1ueda_ |
|
not modelled |
100.0 |
27 |
Fold:Cytochrome P450
Superfamily:Cytochrome P450
Family:Cytochrome P450 |
|
|
| 51 | c4e2pA_ |
|
not modelled |
100.0 |
32 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 107b1 (p450cviib1);
PDBTitle: crystal structure of a post-tailoring hydroxylase (hmtn) involved in2 the himastatin biosynthesis
|
|
|
| 52 | d1s1fa_ |
|
not modelled |
100.0 |
31 |
Fold:Cytochrome P450
Superfamily:Cytochrome P450
Family:Cytochrome P450 |
|
|
| 53 | c1t2bA_ |
|
not modelled |
100.0 |
21 |
PDB header:unknown function
Chain: A: PDB Molecule:p450cin;
PDBTitle: crystal structure of cytochrome p450cin complexed with its2 substrate 1,8-cineole
|
|
|
| 54 | c5l1sA_ |
|
not modelled |
100.0 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pentalenolactone synthase;
PDBTitle: x-ray structure of f232l mutant of cytochrome p450 pntm with2 pentalenolactone f
|
|
|
| 55 | c2jjoA_ |
|
not modelled |
100.0 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 113a1;
PDBTitle: structure of cytochrome p450 eryk in complex with its2 natural substrate erd
|
|
|
| 56 | c3nv6A_ |
|
not modelled |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450;
PDBTitle: crystal structure of camphor-bound cyp101d2
|
|
|
| 57 | c4dxyA_ |
|
not modelled |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450;
PDBTitle: crystal structures of cyp101d2 y96a mutant
|
|
|
| 58 | c4xe3B_ |
|
not modelled |
100.0 |
31 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cytochrome p-450;
PDBTitle: olep, the cytochrome p450 epoxidase from streptomyces antibioticus2 involved in oleandomycin biosynthesis: functional analysis and3 crystallographic structure in complex with clotrimazole.
|
|
|
| 59 | c3tywC_ |
|
not modelled |
100.0 |
29 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative cytochrome p450;
PDBTitle: crystal structure of cyp105n1 from streptomyces coelicolor a3(2)
|
|
|
| 60 | c4yt3B_ |
|
not modelled |
100.0 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cytochrome p450(meg);
PDBTitle: cyp106a2
|
|
|
| 61 | c5y1iA_ |
|
not modelled |
100.0 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450;
PDBTitle: the crystal structure of gfsf
|
|
|
| 62 | c3ivyA_ |
|
not modelled |
100.0 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 cyp125;
PDBTitle: crystal structure of mycobacterium tuberculosis cytochrome p4502 cyp125, p212121 crystal form
|
|
|
| 63 | c6b11B_ |
|
not modelled |
100.0 |
30 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:20-oxo-5-o-mycaminosyltylactone 23-monooxygenase;
PDBTitle: tylhi in complex with native substrate 23-deoxy-5-o-mycaminosyl-2 tylonolide (23-dmtl)
|
|
|
| 64 | c6gmfA_ |
|
not modelled |
100.0 |
32 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative cytochrome p450 hydroxylase;
PDBTitle: structure of cytochrome p450 cyp109q5 from chondromyces apiculatus
|
|
|
| 65 | c5kyoD_ |
|
not modelled |
100.0 |
25 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:cyp101j2;
PDBTitle: crystal structure of cyp101j2
|
|
|
| 66 | c3wecA_ |
|
not modelled |
100.0 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450;
PDBTitle: structure of p450 raua (cyp1050a1) complexed with a biosynthetic2 intermediate of aurachin re
|
|
|
| 67 | c4tvfA_ |
|
not modelled |
100.0 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxyb;
PDBTitle: oxyb from actinoplanes teichomyceticus
|
|
|
| 68 | c4ubsA_ |
|
not modelled |
100.0 |
35 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pentalenic acid synthase;
PDBTitle: the crystal structure of cytochrome p450 105d7 from streptomyces2 avermitilis in complex with diclofenac
|
|
|
| 69 | c2c6hB_ |
|
not modelled |
100.0 |
35 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cytochrome p450 monooxygenase;
PDBTitle: crystal structure of yc-17-bound cytochrome p450 pikc2 (cyp107l1)
|
|
|
| 70 | c4yzrA_ |
|
not modelled |
100.0 |
34 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:polyketide biosynthesis cytochrome p450 pkss;
PDBTitle: bacillus subtilis 168 bacillaene polyketide synthase (pks) cytochrome2 p450 pkss
|
|
|
| 71 | c3o1aA_ |
|
not modelled |
100.0 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxy protein;
PDBTitle: structure of oxye (cyp165d3), a cytochrome p450 involved in2 teicoplanin biosynthesis
|
|
|
| 72 | c2dkkA_ |
|
not modelled |
100.0 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450;
PDBTitle: structure/function studies of cytochrome p450 158a1 from streptomyces2 coelicolor a3(2)
|
|
|
| 73 | c6f0bA_ |
|
not modelled |
100.0 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 monooxygenase;
PDBTitle: cytochrome p450 txtc employs substrate conformational switching for2 sequential aliphatic and aromatic thaxtomin hydroxylation
|
|
|
| 74 | c6g5qA_ |
|
not modelled |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450;
PDBTitle: the structure of a carbohydrate active p450
|
|
|
| 75 | c3ofuE_ |
|
not modelled |
100.0 |
25 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:cytochrome p450;
PDBTitle: crystal structure of cytochrome p450 cyp101c1
|
|
|
| 76 | c4wpzA_ |
|
not modelled |
100.0 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450;
PDBTitle: crystal structure of cytochrome p450 cyp107w1 from streptomyces2 avermitilis
|
|
|
| 77 | c5z9jA_ |
|
not modelled |
100.0 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative p450-like enzyme;
PDBTitle: identification of the functions of unusual cytochrome p450-like2 monooxygenases involved in microbial secondary metablism
|
|
|
| 78 | d1jfba_ |
|
not modelled |
100.0 |
25 |
Fold:Cytochrome P450
Superfamily:Cytochrome P450
Family:Cytochrome P450 |
|
|
| 79 | c6m7lA_ |
|
not modelled |
100.0 |
33 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:putative cytochrome p450 hydroxylase;
PDBTitle: complex of oxya with the x-domain from gpa biosynthesis
|
|
|
| 80 | c2y46B_ |
|
not modelled |
100.0 |
32 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:p-450-like protein;
PDBTitle: structure of the mixed-function p450 mycg in complex with mycinamicin2 iv in c 2 2 21 space group
|
|
|
| 81 | c5ofqD_ |
|
not modelled |
100.0 |
29 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:cytochrome p450;
PDBTitle: crystal structure of substrate-free cyp109a2 from bacillus megaterium
|
|
|
| 82 | c5l90B_ |
|
not modelled |
100.0 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cytochrome p450;
PDBTitle: the crystal structure of substrate-free cyp109e1 from bacillus2 megaterium at 2.55 angstrom resolution
|
|
|
| 83 | c5hiwA_ |
|
not modelled |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 cyp260b1;
PDBTitle: sorangium cellulosum so ce56 cytochrome p450 260b1
|
|
|
| 84 | c4oqrA_ |
|
not modelled |
100.0 |
30 |
PDB header:oxidoreductase/oxidoreductase inhibitor
Chain: A: PDB Molecule:cyp105as1;
PDBTitle: structure of a cyp105as1 mutant in complex with compactin
|
|
|
| 85 | c4jbtB_ |
|
not modelled |
100.0 |
32 |
PDB header:oxidoreductase/substrate
Chain: B: PDB Molecule:cytochrome p450 monooxygenase;
PDBTitle: the 2.2 a crystal structure of cyp154c5 from nocardia farcinica in2 complex with androstenedione
|
|
|
| 86 | d1n40a_ |
|
not modelled |
100.0 |
24 |
Fold:Cytochrome P450
Superfamily:Cytochrome P450
Family:Cytochrome P450 |
|
|
| 87 | c5nwsA_ |
|
not modelled |
100.0 |
33 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:saacmm;
PDBTitle: crystal structure of saacmm involved in actinomycin biosynthesis
|
|
|
| 88 | d1odoa_ |
|
not modelled |
100.0 |
32 |
Fold:Cytochrome P450
Superfamily:Cytochrome P450
Family:Cytochrome P450 |
|
|
| 89 | c4ggvA_ |
|
not modelled |
100.0 |
31 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 superfamily protein;
PDBTitle: crystal structure of hmtt involved in himastatin biosynthesis
|
|
|
| 90 | c3oo3A_ |
|
not modelled |
100.0 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxy protein;
PDBTitle: crystal structure of the orf6* (cyp165d3) monooxygenase involved in2 teicoplanin biosynthesis
|
|
|
| 91 | c4rm4A_ |
|
not modelled |
100.0 |
31 |
PDB header:electron transport
Chain: A: PDB Molecule:cytochrome p450;
PDBTitle: the crystal structure of the versatile cytochrome p450 enzyme cyp109b12 from bacillus subtilis
|
|
|
| 92 | d1io7a_ |
|
not modelled |
100.0 |
27 |
Fold:Cytochrome P450
Superfamily:Cytochrome P450
Family:Cytochrome P450 |
|
|
| 93 | c5cjeA_ |
|
not modelled |
100.0 |
37 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 hydroxylase;
PDBTitle: structure of cyp107l2
|
|
|
| 94 | c5vwsA_ |
|
not modelled |
100.0 |
32 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450;
PDBTitle: ligand free structure of cytochrome p450 tbtj1
|
|
|
| 95 | c5ysmA_ |
|
not modelled |
100.0 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450;
PDBTitle: crystal structure analysis of rif16
|
|
|
| 96 | d1ue8a_ |
|
not modelled |
100.0 |
26 |
Fold:Cytochrome P450
Superfamily:Cytochrome P450
Family:Cytochrome P450 |
|
|
| 97 | c6hqwB_ |
|
not modelled |
100.0 |
30 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cytochrome p450;
PDBTitle: cytochrome p450-153 from novosphingobium aromaticivorans
|
|
|
| 98 | c4z5qA_ |
|
not modelled |
100.0 |
32 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 hydroxylase;
PDBTitle: crystal structure of the lnmz cytochrome p450 hydroxylase from the2 leinamycin biosynthetic pathway of streptomyces atroolivaceus s-1403 at 1.8 a resolution
|
|
|
| 99 | c4l54A_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:terminal olefin-forming fatty acid decarboxylase;
PDBTitle: structure of cytochrome p450 olet, ligand-free
|
|
|
| 100 | c6fyjA_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fatty-acid peroxygenase;
PDBTitle: cytochrome p450 peroxygenase cyp152k6 in complex with myristic acid
|
|
|
| 101 | c3awmA_ |
|
not modelled |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fatty acid alpha-hydroxylase;
PDBTitle: cytochrome p450sp alpha (cyp152b1) wild-type with palmitic acid
|
|
|
| 102 | c2x2nB_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:lanosterol 14-alpha-demethylase;
PDBTitle: x-ray structure of cyp51 from trypanosoma brucei in complex2 with posaconazole in two different conformations
|
|
|
| 103 | d1izoa_ |
|
not modelled |
100.0 |
14 |
Fold:Cytochrome P450
Superfamily:Cytochrome P450
Family:Cytochrome P450 |
|
|
| 104 | c2rfcB_ |
|
not modelled |
100.0 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cytochrome p450;
PDBTitle: ligand bound (4-phenylimidazole) crystal structure of a2 cytochrome p450 from the thermoacidophilic archaeon3 picrophilus torridus
|
|
|
| 105 | c6a18A_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 90b1;
PDBTitle: crystal structure of cyp90b1 in complex with 1,6-hexandiol
|
|
|
| 106 | c5yhjB_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cytochrome p450;
PDBTitle: cytochrome p450ex alpha (cyp152n1) wild-type with myristic acid
|
|
|
| 107 | c2iagA_ |
|
not modelled |
100.0 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:prostacyclin synthase;
PDBTitle: crystal structure of human prostacyclin synthase
|
|
|
| 108 | c4lxjA_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:lanosterol 14-alpha demethylase;
PDBTitle: saccharomyces cerevisiae lanosterol 14-alpha demethylase with2 lanosterol bound
|
|
|
| 109 | c3juvA_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:lanosterol 14-alpha demethylase;
PDBTitle: crystal structure of human lanosterol 14alpha-demethylase (cyp51)
|
|
|
| 110 | c3na0B_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase, electron transport
Chain: B: PDB Molecule:cholesterol side-chain cleavage enzyme, mitochondrial;
PDBTitle: crystal structure of human cyp11a1 in complex with 20,22-2 dihydroxycholesterol
|
|
|
| 111 | c3e4eA_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 2e1;
PDBTitle: human cytochrome p450 2e1 in complex with the inhibitor 4-2 methylpyrazole
|
|
|
| 112 | c3g1qC_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:sterol 14-alpha-demethylase;
PDBTitle: crystal structure of sterol 14-alpha demethylase (cyp51) from2 trypanosoma brucei in ligand free state
|
|
|
| 113 | c2ve3A_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative cytochrome p450 120;
PDBTitle: retinoic acid bound cyanobacterial cyp120a1
|
|
|
| 114 | c6mcwA_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 51;
PDBTitle: crystal structure of the p450 domain of the cyp51-ferredoxin fusion2 protein from methylococcus capsulatus, complex with the detergent3 anapoe-x-114
|
|
|
| 115 | c5fsaA_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cyp51 variant1;
PDBTitle: crystal structure of sterol 14-alpha demethylase (cyp51) from a2 pathogenic yeast candida albicans in complex with the antifungal drug3 posaconazole
|
|
|
| 116 | c2rchA_ |
|
not modelled |
100.0 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:cytochrome p450 74a;
PDBTitle: crystal structure of arabidopsis thaliana allene oxide synthase (aos,2 cytochrome p450 74a, cyp74a) complexed with 13(s)-hod at 1.85 a3 resolution
|
|
|
| 117 | d1tqna_ |
|
not modelled |
100.0 |
19 |
Fold:Cytochrome P450
Superfamily:Cytochrome P450
Family:Cytochrome P450 |
|
|
| 118 | d1r9oa_ |
|
not modelled |
100.0 |
16 |
Fold:Cytochrome P450
Superfamily:Cytochrome P450
Family:Cytochrome P450 |
|
|
| 119 | c5t6qA_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 4b1;
PDBTitle: structure of cytochrome p450 4b1 (cyp4b1) complexed with octane: an n-2 alkane and fatty acid omega-hydroxylase with a covalently bound heme
|
|
|
| 120 | c2q9fA_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome p450 46a1;
PDBTitle: crystal structure of human cytochrome p450 46a1 in complex with2 cholesterol-3-sulphate
|
|
|