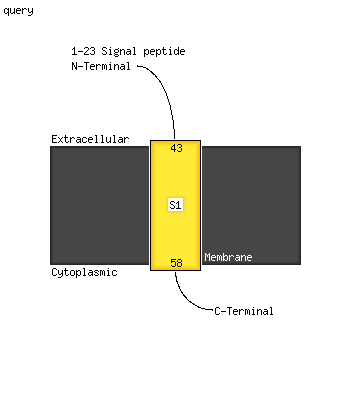
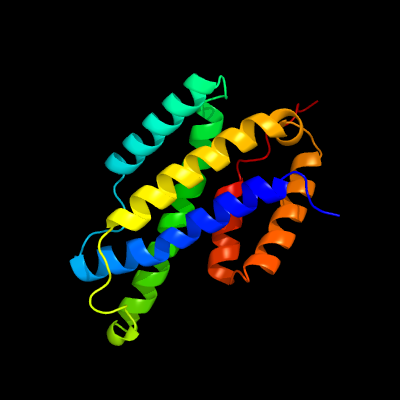
| 1 | d2fp1a1
|
|
 |
100.0 |
100 |
Fold:Chorismate mutase II
Superfamily:Chorismate mutase II
Family:Secreted chorismate mutase-like |
|
|
|
| 2 | c5ts9H_
|
|
 |
100.0 |
25 |
PDB header:isomerase
Chain: H: PDB Molecule:chorismate mutase;
PDBTitle: crystal structure of chorismate mutase from burkholderia phymatum
|
|
|
|
| 3 | c4oj7E_
|
|
 |
100.0 |
30 |
PDB header:isomerase
Chain: E: PDB Molecule:chorismate mutase;
PDBTitle: crystal structure of chorismate mutase from burkholderia thailandensis
|
|
|
|
| 4 | c2gbbA_
|
|
 |
100.0 |
24 |
PDB header:isomerase
Chain: A: PDB Molecule:putative chorismate mutase;
PDBTitle: crystal structure of secreted chorismate mutase from yersinia pestis
|
|
|
|
| 5 | d1ecma_
|
|
 |
99.8 |
25 |
Fold:Chorismate mutase II
Superfamily:Chorismate mutase II
Family:Dimeric chorismate mutase |
|
|
|
| 6 | c3rmiA_
|
|
 |
99.7 |
23 |
PDB header:isomerase
Chain: A: PDB Molecule:chorismate mutase protein;
PDBTitle: crystal structure of chorismate mutase from bartonella henselae str.2 houston-1 in complex with malate
|
|
|
|
| 7 | d2gtvx1
|
|
 |
99.7 |
21 |
Fold:Chorismate mutase II
Superfamily:Chorismate mutase II
Family:monomeric chorismate mutase |
|
|
|
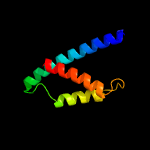
| 8 | c5go2D_
|
|
 |
99.7 |
14 |
PDB header:isomerase
Chain: D: PDB Molecule:protein aroa(g);
PDBTitle: crystal structure of chorismate mutase like domain of bifunctional2 dahp synthase of bacillus subtilis in complex with citrate
|
|
|
|
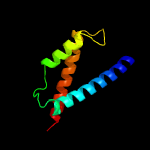
| 9 | c6al9A_
|
|
 |
99.7 |
23 |
PDB header:isomerase
Chain: A: PDB Molecule:chorismate mutase;
PDBTitle: crystal structure of chorismate mutase from helicobacter pylori in2 complex with prephenate
|
|
|
|
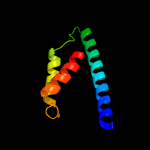
| 10 | d2d8da1
|
|
 |
99.6 |
19 |
Fold:Chorismate mutase II
Superfamily:Chorismate mutase II
Family:Dimeric chorismate mutase |
|
|
|
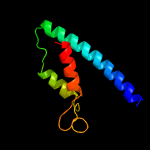
| 11 | d1ybza1
|
|
 |
99.5 |
25 |
Fold:Chorismate mutase II
Superfamily:Chorismate mutase II
Family:Dimeric chorismate mutase |
|
|
|
| 12 | d2h9da1
|
|
 |
99.4 |
21 |
Fold:Chorismate mutase II
Superfamily:Chorismate mutase II
Family:Dimeric chorismate mutase |
|
|
|
| 13 | c3nvtA_
|
|
 |
99.4 |
17 |
PDB header:transferase/isomerase
Chain: A: PDB Molecule:3-deoxy-d-arabino-heptulosonate 7-phosphate synthase;
PDBTitle: 1.95 angstrom crystal structure of a bifunctional 3-deoxy-7-2 phosphoheptulonate synthase/chorismate mutase (aroa) from listeria3 monocytogenes egd-e
|
|
|
|
| 14 | c2qbvA_
|
|
 |
99.2 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:chorismate mutase;
PDBTitle: crystal structure of intracellular chorismate mutase from2 mycobacterium tuberculosis
|
|
|
|
| 15 | c5hubA_
|
|
 |
99.1 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:chorismate mutase;
PDBTitle: high-resolution structure of chorismate mutase from corynebacterium2 glutamicum
|
|
|
|
| 16 | c5w6yB_
|
|
 |
97.5 |
17 |
PDB header:biosynthetic protein,isomerase
Chain: B: PDB Molecule:chorismate mutase;
PDBTitle: physcomitrella patens chorismate mutase
|
|
|
|
| 17 | d5csma_
|
|
 |
97.5 |
24 |
Fold:Chorismate mutase II
Superfamily:Chorismate mutase II
Family:Allosteric chorismate mutase |
|
|
|
| 18 | c4ppuA_
|
|
 |
97.3 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:chorismate mutase 1, chloroplastic;
PDBTitle: crystal structure of atcm1 with tyrosine bound in allosteric site
|
|
|
|
| 19 | c6h3pB_
|
|
 |
97.2 |
16 |
PDB header:plant protein
Chain: B: PDB Molecule:chorismate mutase;
PDBTitle: crystal structure of the cytoplasmic chorismate mutase from zea mays
|
|
|
|
| 20 | c6fpgG_
|
|
 |
96.5 |
25 |
PDB header:cell invasion
Chain: G: PDB Molecule:chromosome 16, whole genome shotgun sequence;
PDBTitle: structure of the ustilago maydis chorismate mutase 1 in complex with a2 zea mays kiwellin
|
|
|
|
| 21 | d1dwka1 |
|
not modelled |
69.9 |
25 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:Cyanase N-terminal domain |
|
|
| 22 | c2iv1J_ |
|
not modelled |
51.2 |
25 |
PDB header:lyase
Chain: J: PDB Molecule:cyanate hydratase;
PDBTitle: site directed mutagenesis of key residues involved in the catalytic2 mechanism of cyanase
|
|
|
| 23 | c5kpeA_ |
|
not modelled |
49.7 |
29 |
PDB header:de novo protein
Chain: A: PDB Molecule:de novo beta sheet design protein or664;
PDBTitle: solution nmr structure of denovo beta sheet design protein, northeast2 structural genomics consortium (nesg) target or664
|
|
|
| 24 | d1gpja3 |
|
not modelled |
31.9 |
14 |
Fold:Ferredoxin-like
Superfamily:Glutamyl tRNA-reductase catalytic, N-terminal domain
Family:Glutamyl tRNA-reductase catalytic, N-terminal domain |
|
|
| 25 | d1myla_ |
|
not modelled |
28.8 |
6 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:Arc/Mnt-like phage repressors |
|
|
| 26 | c4l8tA_ |
|
not modelled |
28.8 |
10 |
PDB header:protein transport
Chain: A: PDB Molecule:unconventional myosin-vc;
PDBTitle: structure of the cargo binding domain from human myosin vc
|
|
|
| 27 | c3n1bA_ |
|
not modelled |
27.7 |
10 |
PDB header:transport protein
Chain: A: PDB Molecule:vacuolar protein sorting-associated protein 54;
PDBTitle: c-terminal domain of vps54 subunit of the garp complex
|
|
|
| 28 | c2ev2B_ |
|
not modelled |
27.3 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:hypothetical protein rv1264/mt1302;
PDBTitle: structure of rv1264n, the regulatory domain of the mycobacterial2 adenylyl cylcase rv1264, at ph 8.5
|
|
|
| 29 | d1icha_ |
|
not modelled |
27.1 |
10 |
Fold:DEATH domain
Superfamily:DEATH domain
Family:DEATH domain, DD |
|
|
| 30 | c1ichA_ |
|
not modelled |
27.1 |
10 |
PDB header:apoptosis
Chain: A: PDB Molecule:tumor necrosis factor receptor-1;
PDBTitle: solution structure of the tumor necrosis factor receptor-12 death domain
|
|
|
| 31 | c5lewA_ |
|
not modelled |
26.7 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:dna polymerase iii subunit alpha;
PDBTitle: dna polymerase
|
|
|
| 32 | d1b28a_ |
|
not modelled |
25.7 |
6 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:Arc/Mnt-like phage repressors |
|
|
| 33 | c1y66D_ |
|
not modelled |
25.3 |
17 |
PDB header:de novo protein
Chain: D: PDB Molecule:engrailed homeodomain;
PDBTitle: dioxane contributes to the altered conformation and2 oligomerization state of a designed engrailed homeodomain3 variant
|
|
|
| 34 | c6noyB_ |
|
not modelled |
23.7 |
30 |
PDB header:structural protein
Chain: B: PDB Molecule:maintenance of carboxysome positioning b protein, mcsb;
PDBTitle: structure of cyanothece mcdb
|
|
|
| 35 | c4j5mA_ |
|
not modelled |
23.6 |
13 |
PDB header:protein transport
Chain: A: PDB Molecule:unconventional myosin-vb;
PDBTitle: structure of the cargo binding domain from human myosin vb
|
|
|
| 36 | d1mylb_ |
|
not modelled |
21.5 |
6 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:Arc/Mnt-like phage repressors |
|
|
| 37 | d2ecca1 |
|
not modelled |
19.7 |
15 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
|
|
| 38 | c2g8yB_ |
|
not modelled |
19.7 |
7 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:malate/l-lactate dehydrogenases;
PDBTitle: the structure of a putative malate/lactate dehydrogenase from e. coli.
|
|
|
| 39 | d2p7vb1 |
|
not modelled |
17.8 |
15 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
|
|
| 40 | c4p9gA_ |
|
not modelled |
17.7 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2,4'-dihydroxyacetophenone dioxygenase;
PDBTitle: structure of the 2,4'-dihydroxyacetophenone dioxygenase from2 alcaligenes sp.
|
|
|
| 41 | c4jrbA_ |
|
not modelled |
17.3 |
16 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:green fluorescent protein;
PDBTitle: structure of cockroach allergen bla g 1 tandem repeat as a egfp fusion
|
|
|
| 42 | c2wtgA_ |
|
not modelled |
17.0 |
11 |
PDB header:oxygen transport
Chain: A: PDB Molecule:globin-like protein;
PDBTitle: high resolution 3d structure of c.elegans globin-like2 protein glb-1
|
|
|
| 43 | c4clvB_ |
|
not modelled |
16.7 |
10 |
PDB header:metal binding protein
Chain: B: PDB Molecule:nickel-cobalt-cadmium resistance protein nccx;
PDBTitle: crystal structure of dodecylphosphocholine-solubilized nccx2 from cupriavidus metallidurans 31a
|
|
|
| 44 | c2knaA_ |
|
not modelled |
16.3 |
8 |
PDB header:apoptosis
Chain: A: PDB Molecule:baculoviral iap repeat-containing protein 4;
PDBTitle: solution structure of uba domain of xiap
|
|
|
| 45 | d1ku3a_ |
|
not modelled |
16.0 |
21 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
|
|
| 46 | d2vo9a1 |
|
not modelled |
15.9 |
3 |
Fold:Hedgehog/DD-peptidase
Superfamily:Hedgehog/DD-peptidase
Family:VanY-like |
|
|
| 47 | c1kcfB_ |
|
not modelled |
15.4 |
7 |
PDB header:hydrolase
Chain: B: PDB Molecule:hypothetical 30.2 kd protein c25g10.02 in
PDBTitle: crystal structure of the yeast mitochondrial holliday2 junction resolvase, ydc2
|
|
|
| 48 | c2k23A_ |
|
not modelled |
15.0 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:lipocalin 2;
PDBTitle: solution structure analysis of the rlcn2
|
|
|
| 49 | c5wujA_ |
|
not modelled |
14.7 |
28 |
PDB header:motor protein
Chain: A: PDB Molecule:flagellar m-ring protein;
PDBTitle: crystal structure of flif-flig complex from h. pylori
|
|
|
| 50 | c2hnhA_ |
|
not modelled |
13.7 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:dna polymerase iii alpha subunit;
PDBTitle: crystal structure of the catalytic alpha subunit of e. coli2 replicative dna polymerase iii
|
|
|
| 51 | d1kcfa2 |
|
not modelled |
13.0 |
8 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Mitochondrial resolvase ydc2 catalytic domain |
|
|
| 52 | c4xp8A_ |
|
not modelled |
13.0 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:etga protein;
PDBTitle: structure of etga d60n mutant
|
|
|
| 53 | c2y8pA_ |
|
not modelled |
12.9 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:endo-type membrane-bound lytic murein transglycosylase a;
PDBTitle: crystal structure of an outer membrane-anchored endolytic2 peptidoglycan lytic transglycosylase (mlte) from3 escherichia coli
|
|
|
| 54 | d1cg5b_ |
|
not modelled |
12.8 |
14 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
|
|
| 55 | c6b2zd_ |
|
not modelled |
12.7 |
24 |
PDB header:membrane protein
Chain: D: PDB Molecule:atp synthase subunit c, mitochondrial;
PDBTitle: cryo-em structure of the dimeric fo region of yeast mitochondrial atp2 synthase
|
|
|
| 56 | c2vo9C_ |
|
not modelled |
12.2 |
3 |
PDB header:hydrolase
Chain: C: PDB Molecule:l-alanyl-d-glutamate peptidase;
PDBTitle: crystal structure of the enzymatically active domain of the listeria2 monocytogenes bacteriophage 500 endolysin ply500
|
|
|
| 57 | c1dlcA_ |
|
not modelled |
12.0 |
15 |
PDB header:toxin
Chain: A: PDB Molecule:delta-endotoxin cryiiia;
PDBTitle: crystal structure of insecticidal delta-endotoxin from2 bacillus thuringiensis at 2.5 angstroms resolution
|
|
|
| 58 | c5n07A_ |
|
not modelled |
11.8 |
8 |
PDB header:transcription
Chain: A: PDB Molecule:hth-type transcriptional repressor nsrr;
PDBTitle: structure of the [4fe-4s] form of the no response regulator nsrr
|
|
|
| 59 | c5l7sA_ |
|
not modelled |
11.3 |
12 |
PDB header:signaling protein
Chain: A: PDB Molecule:secreted rxlr effector peptide protein;
PDBTitle: crystal structure of rxlr effector pexrd54 from phytophthora infestans
|
|
|
| 60 | c6bmcA_ |
|
not modelled |
11.2 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:phospho-2-dehydro-3-deoxyheptonate aldolase;
PDBTitle: the structure of a dimeric type ii dah7ps associated with pyocyanin2 biosynthesis in pseudomonas aeruginosa
|
|
|
| 61 | d1wi3a_ |
|
not modelled |
11.2 |
8 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
|
|
| 62 | c5fwlE_ |
|
not modelled |
10.7 |
16 |
PDB header:chaperone
Chain: E: PDB Molecule:hsp90 co-chaperone cdc37;
PDBTitle: atomic cryoem structure of hsp90-cdc37-cdk4 complex
|
|
|
| 63 | d1gbsa_ |
|
not modelled |
10.6 |
6 |
Fold:Lysozyme-like
Superfamily:Lysozyme-like
Family:G-type lysozyme |
|
|
| 64 | d1jhfa1 |
|
not modelled |
10.5 |
8 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:LexA repressor, N-terminal DNA-binding domain |
|
|
| 65 | d1bazb_ |
|
not modelled |
10.4 |
6 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:Arc/Mnt-like phage repressors |
|
|
| 66 | d2ezha_ |
|
not modelled |
10.3 |
17 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Recombinase DNA-binding domain |
|
|
| 67 | d2d5xa1 |
|
not modelled |
10.2 |
10 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
|
|
| 68 | d1ylfa1 |
|
not modelled |
9.9 |
16 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Transcriptional regulator Rrf2 |
|
|
| 69 | c6hsdB_ |
|
not modelled |
9.9 |
21 |
PDB header:transcription
Chain: B: PDB Molecule:rrf2 family transcriptional regulator;
PDBTitle: crystal structure of the oxidized form of the transcription regulator2 rsrr
|
|
|
| 70 | d1ji6a3 |
|
not modelled |
9.9 |
16 |
Fold:Toxins' membrane translocation domains
Superfamily:delta-Endotoxin (insectocide), N-terminal domain
Family:delta-Endotoxin (insectocide), N-terminal domain |
|
|
| 71 | d1mbaa_ |
|
not modelled |
9.7 |
8 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
|
|
| 72 | c3pt8B_ |
|
not modelled |
9.6 |
13 |
PDB header:oxygen transport
Chain: B: PDB Molecule:hemoglobin iii;
PDBTitle: structure of hbii-iii-cn from lucina pectinata at ph 5.0
|
|
|
| 73 | c3gxkB_ |
|
not modelled |
9.6 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:goose-type lysozyme 1;
PDBTitle: the crystal structure of g-type lysozyme from atlantic cod (gadus2 morhua l.) in complex with nag oligomers sheds new light on substrate3 binding and the catalytic mechanism. native structure to 1.9
|
|
|
| 74 | c3mlgB_ |
|
not modelled |
9.5 |
9 |
PDB header:de novo protein
Chain: B: PDB Molecule:2x chimera of helicobacter pylori protein hp0242;
PDBTitle: 2ouf-2x, a designed knotted protein
|
|
|
| 75 | d1xd7a_ |
|
not modelled |
9.4 |
23 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Transcriptional regulator Rrf2 |
|
|
| 76 | c2wy4A_ |
|
not modelled |
9.3 |
12 |
PDB header:oxygen transport
Chain: A: PDB Molecule:single domain haemoglobin;
PDBTitle: structure of bacterial globin from campylobacter jejuni at2 1.35 a resolution
|
|
|
| 77 | c2l0gA_ |
|
not modelled |
9.2 |
27 |
PDB header:protein binding
Chain: A: PDB Molecule:dna polymerase iota;
PDBTitle: solution nmr structure of ubiquitin-binding motif (ubm2) of human2 polymerase iota
|
|
|
| 78 | d1wh5a_ |
|
not modelled |
9.2 |
0 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
|
|
| 79 | c2eq9C_ |
|
not modelled |
9.2 |
25 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:pyruvate dehydrogenase complex, dihydrolipoamide
PDBTitle: crystal structure of lipoamide dehydrogenase from thermus thermophilus2 hb8 with psbdb
|
|
|
| 80 | c2khuA_ |
|
not modelled |
9.0 |
27 |
PDB header:transferase/protein binding
Chain: A: PDB Molecule:immunoglobulin g-binding protein g, dna
PDBTitle: solution structure of the ubiquitin-binding motif of human2 polymerase iota
|
|
|
| 81 | c2dkzA_ |
|
not modelled |
8.8 |
9 |
PDB header:signaling protein
Chain: A: PDB Molecule:hypothetical protein loc64762;
PDBTitle: solution structure of the sam_pnt-domain of the2 hypothetical protein loc64762
|
|
|
| 82 | d1fvpa_ |
|
not modelled |
8.8 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Non-fluorescent flavoprotein (luxF, FP390) |
|
|
| 83 | c2k29A_ |
|
not modelled |
8.8 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:antitoxin relb;
PDBTitle: structure of the dbd domain of e. coli antitoxin relb
|
|
|
| 84 | d1sr9a1 |
|
not modelled |
8.8 |
27 |
Fold:RuvA C-terminal domain-like
Superfamily:post-HMGL domain-like
Family:DmpG/LeuA communication domain-like |
|
|
| 85 | d1it2a_ |
|
not modelled |
8.7 |
8 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
|
|
| 86 | c2gf5A_ |
|
not modelled |
8.7 |
16 |
PDB header:apoptosis
Chain: A: PDB Molecule:fadd protein;
PDBTitle: structure of intact fadd (mort1)
|
|
|
| 87 | c4zvdA_ |
|
not modelled |
8.7 |
10 |
PDB header:signaling protein
Chain: A: PDB Molecule:diguanylate cyclase dosc;
PDBTitle: crystal structure of mid domain of the e. coli dosc - form ii
|
|
|
| 88 | c3d1lB_ |
|
not modelled |
8.6 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative nadp oxidoreductase bf3122;
PDBTitle: crystal structure of putative nadp oxidoreductase bf3122 from2 bacteroides fragilis
|
|
|
| 89 | c3s1jC_ |
|
not modelled |
8.6 |
14 |
PDB header:oxygen transport, oxygen storage
Chain: C: PDB Molecule:hemoglobin-like flavoprotein;
PDBTitle: crystal structure of acetate-bound hell's gate globin i
|
|
|
| 90 | d1yt3a2 |
|
not modelled |
8.6 |
5 |
Fold:SAM domain-like
Superfamily:HRDC-like
Family:RNase D C-terminal domains |
|
|
| 91 | c4u8uW_ |
|
not modelled |
8.5 |
13 |
PDB header:oxygen storage/transport
Chain: W: PDB Molecule:globin d chain;
PDBTitle: the crystallographic structure of the giant hemoglobin from2 glossoscolex paulistus at 3.2 a resolution.
|
|
|
| 92 | c2eq7C_ |
|
not modelled |
8.4 |
13 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:2-oxoglutarate dehydrogenase e2 component;
PDBTitle: crystal structure of lipoamide dehydrogenase from thermus thermophilus2 hb8 with psbdo
|
|
|
| 93 | c3fdqB_ |
|
not modelled |
8.3 |
17 |
PDB header:dna binding protein/dna
Chain: B: PDB Molecule:motility gene repressor mogr;
PDBTitle: recognition of at-rich dna binding sites by the mogr2 repressor
|
|
|
| 94 | d2qfka1 |
|
not modelled |
8.3 |
8 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
|
|
| 95 | c4q2uM_ |
|
not modelled |
8.2 |
6 |
PDB header:toxin/toxin repressor
Chain: M: PDB Molecule:antitoxin dinj;
PDBTitle: crystal structure of the e. coli dinj-yafq toxin-antitoxin complex
|
|
|
| 96 | c2yqfA_ |
|
not modelled |
8.2 |
13 |
PDB header:protein binding
Chain: A: PDB Molecule:ankyrin-1;
PDBTitle: solution structure of the death domain of ankyrin-1
|
|
|
| 97 | c2zs0A_ |
|
not modelled |
8.2 |
13 |
PDB header:oxygen storage, oxygen transport
Chain: A: PDB Molecule:extracellular giant hemoglobin major globin subunit a1;
PDBTitle: structural basis for the heterotropic and homotropic interactions of2 invertebrate giant hemoglobin
|
|
|
| 98 | d1u9pa1 |
|
not modelled |
8.2 |
6 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:Arc/Mnt-like phage repressors |
|
|
| 99 | c1x3kA_ |
|
not modelled |
8.1 |
5 |
PDB header:oxygen storage/transport
Chain: A: PDB Molecule:hemoglobin component v;
PDBTitle: crystal structure of a hemoglobin component (ta-v) from2 tokunagayusurika akamusi
|
|
|