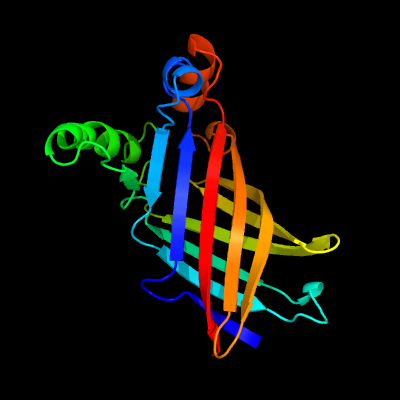
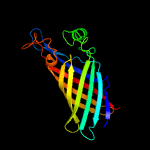
1 d1y0ga_
100.0
14
Fold: Streptavidin-likeSuperfamily: YceI-likeFamily: YceI-like
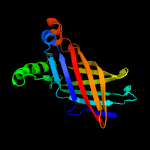
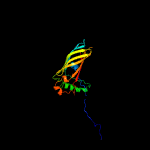
2 d1wuba_
100.0
23
Fold: Streptavidin-likeSuperfamily: YceI-likeFamily: YceI-like
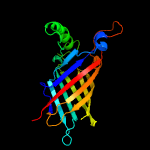
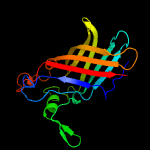
3 c5ixhB_
100.0
25
PDB header: unknown functionChain: B: PDB Molecule: ycei-like domain protein;PDBTitle: crystal structure of burkholderia cenocepacia bcna
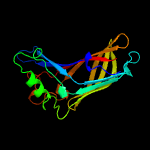
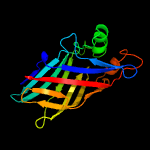
4 c5ixgD_
100.0
16
PDB header: unknown functionChain: D: PDB Molecule: ycei;PDBTitle: crystal structure of burkholderia cenocepacia bcnb
5 c2fgsA_
100.0
16
PDB header: lipid binding proteinChain: A: PDB Molecule: putative periplasmic protein;PDBTitle: crystal structure of campylobacter jejuni ycei protein,2 structural genomics
6 c3hpeB_
100.0
11
PDB header: transport proteinChain: B: PDB Molecule: conserved hypothetical secreted protein;PDBTitle: crystal structure of ycei (hp1286) from helicobacter pylori
7 c2x34A_
100.0
14
PDB header: carbohydrate-binding proteinChain: A: PDB Molecule: cellulose-binding protein, x158;PDBTitle: structure of a polyisoprenoid binding domain from saccharophagus2 degradans implicated in plant cell wall breakdown
8 c3q34A_
100.0
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ycei-like family protein;PDBTitle: the crystal structure of ycei-like family protein from pseudomonas2 syringae
9 c5vtgB_
58.7
14
PDB header: chaperoneChain: B: PDB Molecule: translocation and assembly module subunit tamb;PDBTitle: the structure of tamb963-1138 from escherichia coli reveals a novel2 hydrophobic beta-taco fold
10 c5e6wA_
53.3
10
PDB header: cell adhesionChain: A: PDB Molecule: integrin beta-2;PDBTitle: re-refinement of the crystal structure of the plexin-semaphorin-2 integrin domain/hybrid domain/i-egf1 segment from the human integrin3 b2 subunit
11 c4r80A_
38.8
16
PDB header: de novo proteinChain: A: PDB Molecule: or486;PDBTitle: crystal structure of a de novo designed beta sheet protein, cystatin2 fold, northeast structural genomics consortium (nesg) target or486
12 d1n4ka2
29.2
19
Fold: beta-TrefoilSuperfamily: MIR domainFamily: MIR domain
13 c6fzvA_
20.5
13
PDB header: structural proteinChain: A: PDB Molecule: collagen alpha-1(iii) chain;PDBTitle: crystal structure of the metalloproteinase enhancer pcpe-1 bound to2 the procollagen c propeptide trimer (short)
14 c5k31E_
19.5
13
PDB header: structural proteinChain: E: PDB Molecule: collagen alpha-1(i) chain;PDBTitle: crystal structure of human fibrillar procollagen type i c-propeptide2 homo-trimer
15 d2gqfa2
17.4
15
Fold: HI0933 insert domain-likeSuperfamily: HI0933 insert domain-likeFamily: HI0933 insert domain-like
16 d1pbya4
13.0
27
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Quinohemoprotein amine dehydrogenase A chain, domains 4 and 5
17 c4aejA_
12.7
14
PDB header: structural proteinChain: A: PDB Molecule: collagen alpha-1(iii) chain;PDBTitle: crystal structure of human fibrillar procollagen type iii c-2 propeptide trimer
18 c2vxeA_
10.8
14
PDB header: transcriptionChain: A: PDB Molecule: cg10686-pa;PDBTitle: solution structure of the lsm domain of drosophila2 melanogaster tral (trailer hitch)
19 c1zeqX_
9.8
19
PDB header: metal binding proteinChain: X: PDB Molecule: cation efflux system protein cusf;PDBTitle: 1.5 a structure of apo-cusf residues 6-88 from escherichia2 coli
20 c4qa8A_
8.8
15
PDB header: lipid transportChain: A: PDB Molecule: putative lipoprotein lprf;PDBTitle: crystal structure of lprf from mycobacterium bovis
21 d2vxfa1
not modelled
8.8
16
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: LSM14 N-terminal domain-like
22 c3uc0B_
not modelled
7.6
20
PDB header: viral protein/immune systemChain: B: PDB Molecule: envelope protein;PDBTitle: crystal structure of domain i of the envelope glycoprotein ectodomain2 from dengue virus serotype 4 in complex with the fab fragment of the3 chimpanzee monoclonal antibody 5h2
23 c5ulsA_
not modelled
7.5
53
PDB header: chaperoneChain: A: PDB Molecule: endoplasmin;PDBTitle: structure of grp94 in the active conformation
24 c5tvzA_
not modelled
7.4
12
PDB header: transport proteinChain: A: PDB Molecule: nucleoporin pom152;PDBTitle: solution nmr structure of saccharomyces cerevisiae pom152 ig-like2 repeat, residues 718-820
25 c1y96C_
not modelled
7.3
38
PDB header: rna binding proteinChain: C: PDB Molecule: gem-associated protein 6;PDBTitle: crystal structure of the gemin6/gemin7 heterodimer from the2 human smn complex
26 c5hsbA_
not modelled
7.2
33
PDB header: transcriptionChain: A: PDB Molecule: rna polymerase;PDBTitle: andes virus endonuclease
27 c5tthA_
not modelled
6.7
59
PDB header: chaperoneChain: A: PDB Molecule: c-terminal spycatcher fusion of wildtype zebrafish tnfPDBTitle: heterodimeric spycatcher/spytag-fused zebrafish trap1 in atp/adp-2 hybrid state
28 c4ipeA_
not modelled
6.6
59
PDB header: chaperoneChain: A: PDB Molecule: tnf receptor-associated protein 1;PDBTitle: crystal structure of mitochondrial hsp90 (trap1) with amppnp
29 c2l55A_
not modelled
6.4
24
PDB header: metal binding proteinChain: A: PDB Molecule: silb,silver efflux protein, mfp component of the threePDBTitle: solution structure of the c-terminal domain of silb from cupriavidus2 metallidurans
30 d2icha1
not modelled
6.2
11
Fold: AttH-likeSuperfamily: AttH-likeFamily: AttH-like
31 c3d8mA_
not modelled
6.2
23
PDB header: virus/viral proteinChain: A: PDB Molecule: baseplate protein, receptor binding protein;PDBTitle: crystal structure of a chimeric receptor binding protein from2 lactococcal phages subspecies tp901-1 and p2
32 d1u7ba2
not modelled
5.9
13
Fold: DNA clampSuperfamily: DNA clampFamily: DNA polymerase processivity factor
33 c3da0C_
not modelled
5.6
23
PDB header: viral proteinChain: C: PDB Molecule: cleaved chimeric receptor binding protein fromPDBTitle: crystal structure of a cleaved form of a chimeric receptor binding2 protein from lactococcal phages subspecies tp901-1 and p2
34 d1rwza2
not modelled
5.4
12
Fold: DNA clampSuperfamily: DNA clampFamily: DNA polymerase processivity factor
35 c2qazC_
not modelled
5.3
16
PDB header: hydrolase activatorChain: C: PDB Molecule: sspb protein;PDBTitle: structure of c. crescentus sspb ortholog
36 d1ud9a2
not modelled
5.0
31
Fold: DNA clampSuperfamily: DNA clampFamily: DNA polymerase processivity factor
37 d2nysa1
not modelled
5.0
19
Fold: SspB-likeSuperfamily: SspB-likeFamily: AGR C 3712p-like