| 1 |
|
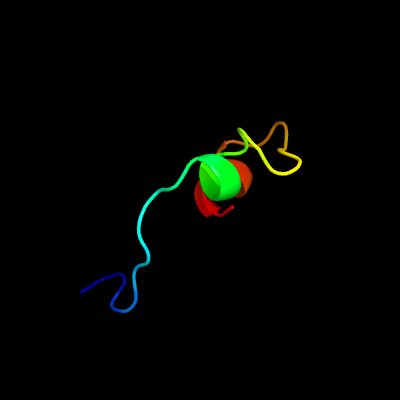
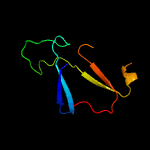
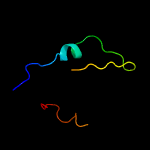
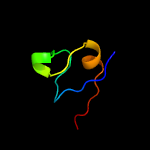
PDB 2ciw chain A domain 1
Region: 75 - 104
Aligned: 28
Modelled: 30
Confidence: 15.1%
Identity: 32%
Fold: EF Hand-like
Superfamily: Cloroperoxidase
Family: Cloroperoxidase
Phyre2
| 2 |
|
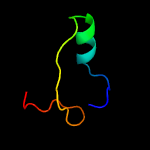
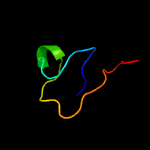
PDB 2yp1 chain A
Region: 67 - 104
Aligned: 34
Modelled: 38
Confidence: 14.7%
Identity: 21%
PDB header:oxidoreductase
Chain: A: PDB Molecule:aromatic peroxygenase;
PDBTitle: crystallization of a 45 kda peroxygenase- peroxidase from2 the mushroom agrocybe aegerita and structure determination3 by sad utilizing only the haem iron
Phyre2
| 3 |
|
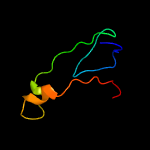
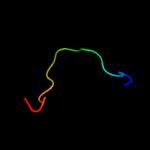
PDB 1mwp chain A
Region: 62 - 85
Aligned: 22
Modelled: 24
Confidence: 14.6%
Identity: 23%
Fold: SRCR-like
Superfamily: A heparin-binding domain
Family: A heparin-binding domain
Phyre2
| 4 |
|
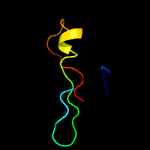
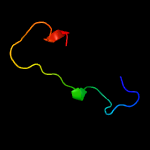
PDB 5td8 chain D
Region: 36 - 101
Aligned: 45
Modelled: 46
Confidence: 13.9%
Identity: 22%
PDB header:replication
Chain: D: PDB Molecule:kinetochore protein spc25;
PDBTitle: crystal structure of an extended dwarf ndc80 complex
Phyre2
| 5 |
|
PDB 2djm chain A
Region: 48 - 81
Aligned: 29
Modelled: 34
Confidence: 13.6%
Identity: 17%
PDB header:sugar binding protein
Chain: A: PDB Molecule:glucoamylase a;
PDBTitle: solution structure of n-terminal starch-binding domain of2 glucoamylase from rhizopus oryzae
Phyre2
| 6 |
|
PDB 3pkw chain A
Region: 50 - 122
Aligned: 70
Modelled: 73
Confidence: 12.1%
Identity: 20%
PDB header:lyase
Chain: A: PDB Molecule:toxoflavin lyase (tfla);
PDBTitle: crystal structure of toxoflavin lyase (tfla) bound to mn(ii)
Phyre2
| 7 |
|
PDB 5fuj chain B
Region: 92 - 104
Aligned: 13
Modelled: 13
Confidence: 11.7%
Identity: 31%
PDB header:oxidoreductase
Chain: B: PDB Molecule:mroupo;
PDBTitle: crystallization of a dimeric heme peroxygenase from the2 fungus marasmius rotula
Phyre2
| 8 |
|
PDB 1sgv chain A domain 1
Region: 77 - 109
Aligned: 22
Modelled: 33
Confidence: 10.2%
Identity: 32%
Fold: PUA domain-like
Superfamily: PUA domain-like
Family: PUA domain
Phyre2
| 9 |
|
PDB 3brd chain A domain 1
Region: 43 - 94
Aligned: 51
Modelled: 52
Confidence: 10.2%
Identity: 27%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: E set domains
Family: NF-kappa-B/REL/DORSAL transcription factors, C-terminal domain
Phyre2
| 10 |
|
PDB 3ktm chain B
Region: 62 - 85
Aligned: 22
Modelled: 24
Confidence: 9.5%
Identity: 23%
PDB header:cell adhesion, signaling protein
Chain: B: PDB Molecule:amyloid beta a4 protein;
PDBTitle: structure of the heparin-induced e1-dimer of the amyloid precursor2 protein (app)
Phyre2
| 11 |
|
PDB 5lc5 chain Q
Region: 47 - 83
Aligned: 32
Modelled: 37
Confidence: 8.9%
Identity: 22%
PDB header:oxidoreductase
Chain: Q: PDB Molecule:nadh dehydrogenase [ubiquinone] iron-sulfur protein 4,
PDBTitle: structure of mammalian respiratory complex i, class2
Phyre2
| 12 |
|
PDB 5wrg chain B
Region: 26 - 65
Aligned: 40
Modelled: 40
Confidence: 7.9%
Identity: 28%
PDB header:virus like particle
Chain: B: PDB Molecule:spike glycoprotein;
PDBTitle: sars-cov spike glycoprotein
Phyre2
| 13 |
|
PDB 1en4 chain C
Region: 90 - 121
Aligned: 30
Modelled: 32
Confidence: 6.7%
Identity: 23%
PDB header:oxidoreductase
Chain: C: PDB Molecule:manganese superoxide dismutase;
PDBTitle: crystal structure analysis of the e. coli manganese superoxide2 dismutase q146h mutant
Phyre2
| 14 |
|
PDB 1qfn chain B
Region: 30 - 46
Aligned: 17
Modelled: 17
Confidence: 6.6%
Identity: 35%
PDB header:electron transport/oxidoreductase
Chain: B: PDB Molecule:protein (ribonucleoside-diphosphate reductase 1);
PDBTitle: glutaredoxin-1-ribonucleotide reductase b1 mixed disulfide2 bond
Phyre2
| 15 |
|
PDB 2hig chain A
Region: 76 - 103
Aligned: 28
Modelled: 28
Confidence: 6.5%
Identity: 18%
PDB header:transferase
Chain: A: PDB Molecule:6-phospho-1-fructokinase;
PDBTitle: crystal structure of phosphofructokinase apoenzyme from trypanosoma2 brucei.
Phyre2
| 16 |
|
PDB 5a4h chain A
Region: 116 - 124
Aligned: 9
Modelled: 9
Confidence: 6.4%
Identity: 56%
PDB header:transferase
Chain: A: PDB Molecule:1-acylglycerol-3-phosphate o-acyltransferase abhd5;
PDBTitle: solution structure of the lipid droplet anchoring peptide2 of cgi-58 bound to dpc micelles
Phyre2
| 17 |
|
PDB 2cj0 chain A
Region: 92 - 104
Aligned: 13
Modelled: 13
Confidence: 5.9%
Identity: 46%
PDB header:oxidoreductase
Chain: A: PDB Molecule:chloroperoxidase;
PDBTitle: chloroperoxidase complexed with nitrate
Phyre2
| 18 |
|
PDB 5aj3 chain D
Region: 48 - 63
Aligned: 16
Modelled: 16
Confidence: 5.8%
Identity: 13%
PDB header:ribosome
Chain: D: PDB Molecule:
PDBTitle: structure of the small subunit of the mammalian mitoribosome
Phyre2
| 19 |
|
PDB 1ix9 chain A domain 2
Region: 87 - 121
Aligned: 33
Modelled: 35
Confidence: 5.3%
Identity: 24%
Fold: Fe,Mn superoxide dismutase (SOD), C-terminal domain
Superfamily: Fe,Mn superoxide dismutase (SOD), C-terminal domain
Family: Fe,Mn superoxide dismutase (SOD), C-terminal domain
Phyre2