1 c2wvrB_
33.0
21
PDB header: replicationChain: B: PDB Molecule: geminin;PDBTitle: human cdt1:geminin complex
2 d2h8pc1
16.9
25
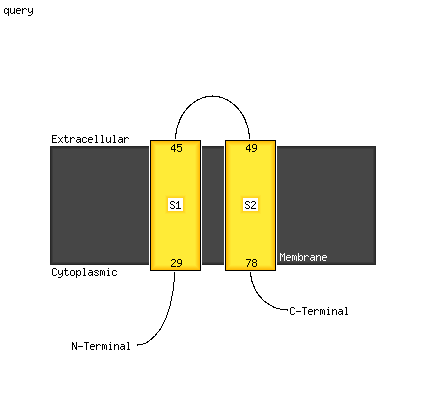
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels
3 c5z58w_
15.8
67
PDB header: splicingChain: W: PDB Molecule: PDBTitle: cryo-em structure of a human activated spliceosome (early bact) at 4.92 angstrom.
4 c5oqtC_
13.5
50
PDB header: transport proteinChain: C: PDB Molecule: uncharacterized protein ynem;PDBTitle: crystal structure of a bacterial cationic amino acid transporter (cat)2 homologue
5 c1hk7A_
13.4
58
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein hsp82;PDBTitle: middle domain of hsp90
6 c6g90T_
13.2
50
PDB header: splicingChain: T: PDB Molecule: pre-mrna-splicing factor prp9;PDBTitle: prespliceosome structure provides insight into spliceosome assembly2 and regulation (map a2)
7 c1y6zA_
13.2
50
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: heat shock protein, putative;PDBTitle: middle domain of plasmodium falciparum putative heat shock protein2 pf14_0417
8 c5jygA_
13.1
43
PDB header: structural proteinChain: A: PDB Molecule: actin-like atpase;PDBTitle: cryo-em structure of the mamk filament at 6.5 a
9 c6f34C_
12.4
50
PDB header: membrane proteinChain: C: PDB Molecule: mgts;PDBTitle: crystal structure of a bacterial cationic amino acid transporter (cat)2 homologue bound to arginine.
10 c2qw6A_
12.1
33
PDB header: hydrolaseChain: A: PDB Molecule: aaa atpase, central region;PDBTitle: crystal structure of the c-terminal domain of an aaa atpase from2 enterococcus faecium do
11 d2qw6a1
12.1
33
Fold: post-AAA+ oligomerization domain-likeSuperfamily: post-AAA+ oligomerization domain-likeFamily: MgsA/YrvN C-terminal domain-like
12 d1usua_
11.6
50
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Hsp90 middle domain
13 c2ww9O_
11.5
50
PDB header: ribosomeChain: O: PDB Molecule: 60s ribosomal protein l39;PDBTitle: cryo-em structure of the active yeast ssh1 complex bound to the yeast2 80s ribosome
14 c2o1tB_
11.4
33
PDB header: chaperoneChain: B: PDB Molecule: endoplasmin;PDBTitle: structure of middle plus c-terminal domains (m+c) of grp94
15 d1dxsa_
11.1
62
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: SAM (sterile alpha motif) domain
16 c4xb6D_
10.7
35
PDB header: transferaseChain: D: PDB Molecule: alpha-d-ribose 1-methylphosphonate 5-phosphate c-p lyase;PDBTitle: structure of the e. coli c-p lyase core complex
17 c3pryA_
9.8
33
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein hsp 90-beta;PDBTitle: crystal structure of the middle domain of human hsp90-beta refined at2 2.3 a resolution
18 c2xuvB_
9.7
54
PDB header: unknown functionChain: B: PDB Molecule: hdeb;PDBTitle: the structure of hdeb
19 d1rg6a_
9.5
26
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: SAM (sterile alpha motif) domain
20 c2zkr3_
9.3
63
PDB header: ribosomal protein/rnaChain: 3: PDB Molecule: 60s ribosomal protein l39e;PDBTitle: structure of a mammalian ribosomal 60s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
21 c2gq0B_
not modelled
8.6
50
PDB header: chaperone, hydrolaseChain: B: PDB Molecule: chaperone protein htpg;PDBTitle: crystal structure of the middle domain of htpg, the e. coli hsp90
22 c3q6nF_
not modelled
8.5
42
PDB header: chaperoneChain: F: PDB Molecule: heat shock protein hsp 90-alpha;PDBTitle: crystal structure of human mc-hsp90 in p21 space group
23 c5xfmD_
not modelled
8.5
50
PDB header: hydrolaseChain: D: PDB Molecule: alpha-glucosidase;PDBTitle: crystal structure of beta-arabinopyranosidase
24 c2la3A_
not modelled
8.1
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: the nmr structure of the protein np_344798.1 reveals a cca-adding2 enzyme head domain
25 c2cgeD_
not modelled
8.0
58
PDB header: chaperoneChain: D: PDB Molecule: atp-dependent molecular chaperone hsp82;PDBTitle: crystal structure of an hsp90-sba1 closed chaperone complex
26 c3hjcA_
not modelled
7.9
50
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein 83-1;PDBTitle: crystal structure of the carboxy-terminal domain of hsp90 from2 leishmania major, lmjf33.0312
27 c3a24A_
not modelled
6.9
50
PDB header: hydrolaseChain: A: PDB Molecule: alpha-galactosidase;PDBTitle: crystal structure of bt1871 retaining glycosidase
28 c2n90A_
not modelled
6.8
67
PDB header: transferaseChain: A: PDB Molecule: high affinity nerve growth factor receptor;PDBTitle: trka transmembrane domain nmr structure in dpc micelles
29 c2n90B_
not modelled
6.8
67
PDB header: transferaseChain: B: PDB Molecule: high affinity nerve growth factor receptor;PDBTitle: trka transmembrane domain nmr structure in dpc micelles
30 c3zf7q_
not modelled
6.7
40
PDB header: ribosomeChain: Q: PDB Molecule: ribosomal protein l15;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
31 c3r9jD_
not modelled
6.6
18
PDB header: cell cycle,hydrolase/cell cycleChain: D: PDB Molecule: cell division topological specificity factor;PDBTitle: 4.3a resolution structure of a mind-mine(i24n) protein complex
32 c3izso_
not modelled
6.4
40
PDB header: ribosomeChain: O: PDB Molecule: 60s ribosomal protein rpl28 (l15p);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
33 d1xmec1
not modelled
6.3
23
Fold: Single transmembrane helixSuperfamily: Bacterial ba3 type cytochrome c oxidase subunit IIaFamily: Bacterial ba3 type cytochrome c oxidase subunit IIa
34 c3bvdC_
not modelled
6.3
23
PDB header: oxidoreductaseChain: C: PDB Molecule: cytochrome c oxidase polypeptide 2a;PDBTitle: structure of surface-engineered cytochrome ba3 oxidase from thermus2 thermophilus under xenon pressure, 100psi 5min
35 c6adqP_
not modelled
6.2
23
PDB header: electron transportChain: P: PDB Molecule: prokaryotic respiratory supercomplex associate factor 1PDBTitle: respiratory complex ciii2civ2sod2 from mycobacterium smegmatis
36 c2iopD_
not modelled
5.9
50
PDB header: chaperoneChain: D: PDB Molecule: chaperone protein htpg;PDBTitle: crystal structure of full-length htpg, the escherichia coli2 hsp90, bound to adp
37 c2wfoA_
not modelled
5.7
43
PDB header: viral proteinChain: A: PDB Molecule: glycoprotein 1;PDBTitle: crystal structure of machupo virus envelope glycoprotein gp1
38 c5tthA_
not modelled
5.6
42
PDB header: chaperoneChain: A: PDB Molecule: c-terminal spycatcher fusion of wildtype zebrafish tnfPDBTitle: heterodimeric spycatcher/spytag-fused zebrafish trap1 in atp/adp-2 hybrid state
39 c2ariA_
not modelled
5.5
50
PDB header: viral proteinChain: A: PDB Molecule: envelope polyprotein gp160;PDBTitle: solution structure of micelle-bound fusion domain of hiv-12 gp41
40 c2wseE_
not modelled
5.1
40
PDB header: photosynthesisChain: E: PDB Molecule: photosystem i reaction center subunit iv a, chloroplastic;PDBTitle: improved model of plant photosystem i