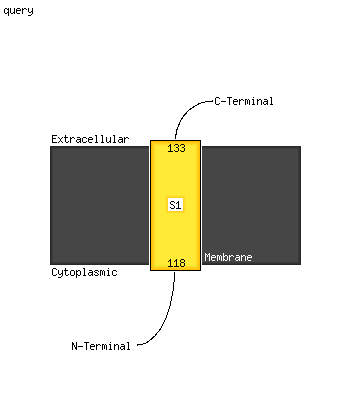
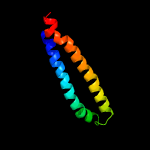
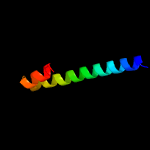
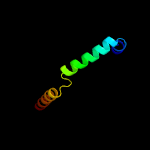
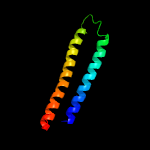
1 c5xfsB_
100.0
51
PDB header: protein transportChain: B: PDB Molecule: ppe family protein ppe15;PDBTitle: crystal structure of pe8-ppe15 in complex with espg5 from m.2 tuberculosis
2 c2g38B_
100.0
34
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: ppe family protein;PDBTitle: a pe/ppe protein complex from mycobacterium tuberculosis
3 d2g38b1
100.0
34
Fold: Ferritin-likeSuperfamily: PE/PPE dimer-likeFamily: PPE
4 c4xy3A_
100.0
19
PDB header: protein transportChain: A: PDB Molecule: esx-1 secretion-associated protein espb;PDBTitle: structure of esx-1 secreted protein espb
5 c4wj2A_
96.7
21
PDB header: unknown functionChain: A: PDB Molecule: antigen mtb48;PDBTitle: mycobacterial protein
6 c4iogD_
95.7
16
PDB header: unknown functionChain: D: PDB Molecule: secreted protein esxb;PDBTitle: the crystal structure of a secreted protein esxb (wild-type, in p212 space group) from bacillus anthracis str. sterne
7 c2vs0B_
95.5
9
PDB header: cell invasionChain: B: PDB Molecule: virulence factor esxa;PDBTitle: structural analysis of homodimeric staphylococcal aureus2 virulence factor esxa
8 c3gvmA_
94.4
11
PDB header: viral proteinChain: A: PDB Molecule: putative uncharacterized protein sag1039;PDBTitle: structure of the homodimeric wxg-100 family protein from streptococcus2 agalactiae
9 c3zbhC_
94.2
16
PDB header: unknown functionChain: C: PDB Molecule: esxa;PDBTitle: geobacillus thermodenitrificans esxa crystal form i
10 d1wa8a1
92.4
15
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like
11 c3jywF_
69.2
43
PDB header: ribosomeChain: F: PDB Molecule: 60s ribosomal protein l7(a);PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
12 c4lwsA_
65.7
19
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
13 d1wa8b1
58.8
16
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like
14 c4i0xA_
47.6
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: esat-6-like protein mab_3112;PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
15 c2vsnB_
45.7
12
PDB header: transferaseChain: B: PDB Molecule: xcogt;PDBTitle: structure and topological arrangement of an o-glcnac2 transferase homolog: insight into molecular control of3 intracellular glycosylation
16 c4lwsB_
37.8
9
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
17 c2kg7B_
31.5
16
PDB header: unknown functionChain: B: PDB Molecule: esat-6-like protein esxh;PDBTitle: structure and features of the complex formed by the tuberculosis2 virulence factors rv0287 and rv0288
18 c4bgoA_
29.0
12
PDB header: hydrolaseChain: A: PDB Molecule: efem m75 peptidase;PDBTitle: structural and functional role of the imelysin-like protein2 efem from pseudomonas syringae pv. syringae and3 implications in bacterial iron transport
19 c5djsA_
24.0
18
PDB header: transferaseChain: A: PDB Molecule: tetratricopeptide tpr_2 repeat protein;PDBTitle: thermobaculum terrenum o-glcnac transferase mutant - k341m
20 c5uz9B_
21.5
21
PDB header: immune system/rnaChain: B: PDB Molecule: crispr-associated protein csy2;PDBTitle: cryo em structure of anti-crisprs, acrf1 and acrf2, bound to type i-f2 crrna-guided crispr surveillance complex
21 c2lyyB_
not modelled
20.5
18
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: nmr structure of the protein nb7890a from shewanella sp
22 d1ui5a2
not modelled
20.0
20
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain
23 c3pe3D_
not modelled
19.9
14
PDB header: transferaseChain: D: PDB Molecule: udp-n-acetylglucosamine--peptide n-PDBTitle: structure of human o-glcnac transferase and its complex with a peptide2 substrate
24 c3at7B_
not modelled
19.9
9
PDB header: structural proteinChain: B: PDB Molecule: alginate-binding flagellin;PDBTitle: crystal structure of bacterial cell-surface alginate-binding protein2 algp7
25 c4lrvL_
not modelled
18.2
40
PDB header: dna binding proteinChain: L: PDB Molecule: dna sulfur modification protein dnde;PDBTitle: crystal structure of dnde from escherichia coli b7a involved in dna2 phosphorothioation modification
26 c3fpjA_
not modelled
17.6
11
PDB header: biosynthetic protein, transferaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of e81q mutant of mtnas in complex with s-2 adenosylmethionine
27 c6ghrF_
not modelled
17.1
53
PDB header: photosynthesisChain: F: PDB Molecule: cp12 polypeptide;PDBTitle: cyanobacterial gapdh with full-length cp12
28 c3lhoA_
not modelled
16.2
28
PDB header: hydrolaseChain: A: PDB Molecule: putative hydrolase;PDBTitle: crystal structure of putative hydrolase (yp_751971.1) from shewanella2 frigidimarina ncimb 400 at 1.80 a resolution
29 c4jzaB_
not modelled
15.9
58
PDB header: hydrolaseChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a legionella phosphoinositide phosphatase:2 insights into lipid metabolism in pathogen host interaction
30 d1fnda2
not modelled
15.9
26
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: Reductases
31 d2a15a1
not modelled
15.1
24
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Ketosteroid isomerase-like
32 d1knca_
not modelled
14.5
19
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: AhpD
33 d1rzoa_
not modelled
13.8
10
Fold: Ribosome inactivating proteins (RIP)Superfamily: Ribosome inactivating proteins (RIP)Family: Plant cytotoxins
34 c3kb4D_
not modelled
13.2
14
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: alr8543 protein;PDBTitle: crystal structure of the alr8543 protein in complex with2 geranylgeranyl monophosphate and magnesium ion from nostoc sp. pcc3 7120, northeast structural genomics consortium target nsr141
35 c4fbaC_
not modelled
13.2
25
PDB header: hydrolaseChain: C: PDB Molecule: protein synthesis inhibitor i;PDBTitle: structure of mutant rip from barley seeds in complex with adenine
36 d1hi9a_
not modelled
12.4
26
Fold: Dipeptide transport proteinSuperfamily: Dipeptide transport proteinFamily: Dipeptide transport protein
37 d2fr1a1
not modelled
12.0
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases
38 d1rp4a_
not modelled
11.6
21
Fold: ERO1-likeSuperfamily: ERO1-likeFamily: ERO1-like
39 c3ahrA_
not modelled
11.1
28
PDB header: oxidoreductaseChain: A: PDB Molecule: ero1-like protein alpha;PDBTitle: inactive human ero1
40 c2pqjC_
not modelled
11.0
24
PDB header: hydrolaseChain: C: PDB Molecule: ribosome-inactivating protein 3;PDBTitle: crystal structure of active ribosome inactivating protein from maize2 (b-32), complex with adenine
41 c4o94B_
not modelled
10.4
15
PDB header: transport proteinChain: B: PDB Molecule: trap dicarboxylate transporter dctp subunit;PDBTitle: crystal structure of a trap periplasmic solute binding protein from2 rhodopseudomonas palustris haa2 (rpb_3329), target efi-510223, with3 bound succinate
42 c3q3hA_
not modelled
10.3
10
PDB header: transferaseChain: A: PDB Molecule: hmw1c-like glycosyltransferase;PDBTitle: crystal structure of the actinobacillus pleuropneumoniae hmw1c2 glycosyltransferase in complex with udp-glc
43 d1uq5a_
not modelled
10.2
10
Fold: Ribosome inactivating proteins (RIP)Superfamily: Ribosome inactivating proteins (RIP)Family: Plant cytotoxins
44 d1f20a2
not modelled
9.9
26
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: NADPH-cytochrome p450 reductase-like
45 c5frgA_
not modelled
9.8
75
PDB header: protein bindingChain: A: PDB Molecule: formin-binding protein 1-like;PDBTitle: the nmr structure of the cdc42-interacting region of toca1
46 d1yf8a1
not modelled
9.7
13
Fold: Ribosome inactivating proteins (RIP)Superfamily: Ribosome inactivating proteins (RIP)Family: Plant cytotoxins
47 c4co6A_
not modelled
9.6
37
PDB header: chaperoneChain: A: PDB Molecule: nucleoprotein;PDBTitle: crystal structure of the nipah virus rna free2 nucleoprotein-phosphoprotein complex
48 c4rqoB_
not modelled
9.6
25
PDB header: lyaseChain: B: PDB Molecule: l-serine dehydratase;PDBTitle: crystal structure of l-serine dehydratase from legionella pneumophila
49 c1trlB_
not modelled
9.5
28
PDB header: hydrolase (metalloprotease)Chain: B: PDB Molecule: thermolysin fragment 255 - 316;PDBTitle: nmr solution structure of the c-terminal fragment 255-3162 of thermolysin: a dimer formed by subunits having the3 native structure
50 d1g03a_
not modelled
9.2
19
Fold: Retrovirus capsid protein, N-terminal core domainSuperfamily: Retrovirus capsid protein, N-terminal core domainFamily: Retrovirus capsid protein, N-terminal core domain
51 d1vefa1
not modelled
9.1
16
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like
52 c1bkvA_
not modelled
9.0
38
PDB header: structural proteinChain: A: PDB Molecule: t3-785;PDBTitle: collagen
53 d1umka2
not modelled
8.9
21
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: Reductases
54 c5ddwD_
not modelled
8.8
22
PDB header: transferaseChain: D: PDB Molecule: crmg;PDBTitle: crystal structure of aminotransferase crmg from actinoalloteichus sp.2 wh1-2216-6 in complex with the pmp external aldimine adduct with3 caerulomycin m
55 d3cx5d1
not modelled
8.6
35
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Cytochrome bc1 domain
56 d1byra_
not modelled
8.6
22
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Nuclease
57 d1oc7a_
not modelled
8.6
14
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycosyl hydrolases family 6, cellulasesFamily: Glycosyl hydrolases family 6, cellulases
58 d1spgb_
not modelled
8.5
29
Fold: Globin-likeSuperfamily: Globin-likeFamily: Globins
59 c1qcrD_
not modelled
8.5
30
PDB header: oxidoreductaseChain: D: PDB Molecule: ubiquinol cytochrome c oxidoreductase;PDBTitle: crystal structure of bovine mitochondrial cytochrome bc12 complex, alpha carbon atoms only
60 c1bkvC_
not modelled
8.3
38
PDB header: structural proteinChain: C: PDB Molecule: t3-785;PDBTitle: collagen
61 c1bkvB_
not modelled
8.3
38
PDB header: structural proteinChain: B: PDB Molecule: t3-785;PDBTitle: collagen
62 c3fyqA_
not modelled
8.3
21
PDB header: cell adhesionChain: A: PDB Molecule: cg6831-pa (talin);PDBTitle: structure of drosophila melanogaster talin ibs2 domain (residues 1981-2 2168)
63 c3vohA_
not modelled
8.3
18
PDB header: hydrolaseChain: A: PDB Molecule: cellobiohydrolase;PDBTitle: cccel6a catalytic domain complexed with cellobiose
64 c3bkhA_
not modelled
8.2
20
PDB header: hydrolaseChain: A: PDB Molecule: lytic transglycosylase;PDBTitle: crystal structure of the bacteriophage phikz lytic2 transglycosylase, gp144
65 d1ppjd1
not modelled
8.1
30
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Cytochrome bc1 domain
66 d1luaa2
not modelled
7.9
36
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Methylene-tetrahydromethanopterin dehydrogenase
67 c1sseB_
not modelled
7.9
18
PDB header: transcription activatorChain: B: PDB Molecule: ap-1 like transcription factor yap1;PDBTitle: solution structure of the oxidized form of the yap1 redox2 domain
68 c2pheC_
not modelled
7.9
31
PDB header: transcriptionChain: C: PDB Molecule: alpha trans-inducing protein;PDBTitle: model for vp16 binding to pc4
69 c6efvA_
not modelled
7.9
28
PDB header: flavoproteinChain: A: PDB Molecule: sulfite reductase [nadph] flavoprotein alpha-component;PDBTitle: the nadph-dependent sulfite reductase flavoprotein adopts an extended2 conformation that is unique to this diflavin reductase
70 c6fhjA_
not modelled
7.9
20
PDB header: hydrolaseChain: A: PDB Molecule: protein,protein;PDBTitle: structural dynamics and catalytic properties of a multi-modular2 xanthanase, native.
71 c6rdf7_
not modelled
7.8
30
PDB header: proton transportChain: 7: PDB Molecule: mitochondrial atp synthase associated protein asa7;PDBTitle: cryoem structure of polytomella f-atp synthase, primary rotary state2 3, monomer-masked refinement
72 c4xjnM_
not modelled
7.7
32
PDB header: viral protein/rnaChain: M: PDB Molecule: nucleocapsid;PDBTitle: structure of the parainfluenza virus 5 nucleocapsid-rna complex: an2 insight into paramyxovirus polymerase activity
73 c5lzkB_
not modelled
7.6
6
PDB header: structural genomicsChain: B: PDB Molecule: protein fam83b;PDBTitle: structure of the domain of unknown function duf1669 from human fam83b
74 c2phgB_
not modelled
7.4
33
PDB header: transcriptionChain: B: PDB Molecule: alpha trans-inducing protein;PDBTitle: model for vp16 binding to tfiib
75 d1dwna_
not modelled
7.4
38
Fold: RNA bacteriophage capsid proteinSuperfamily: RNA bacteriophage capsid proteinFamily: RNA bacteriophage capsid protein
76 c3a64A_
not modelled
7.1
17
PDB header: hydrolaseChain: A: PDB Molecule: cellobiohydrolase;PDBTitle: crystal structure of cccel6c, a glycoside hydrolase family 62 enzyme, from coprinopsis cinerea
77 c5jn6A_
not modelled
7.1
67
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: the nmr solution structure of rpa3313
78 c5cgaC_
not modelled
7.0
33
PDB header: transferaseChain: C: PDB Molecule: hydroxyethylthiazole kinase;PDBTitle: structure of hydroxyethylthiazole kinase thim from staphylococcus2 aureus in complex with substrate analog 2-(1,3,5-trimethyl-1h-3 pyrazole-4-yl)ethanol
79 c5e4vA_
not modelled
7.0
32
PDB header: viral proteinChain: A: PDB Molecule: nucleoprotein,phosphoprotein;PDBTitle: crystal structure of measles n0-p complex
80 c3ktzA_
not modelled
7.0
15
PDB header: hydrolaseChain: A: PDB Molecule: ribosome-inactivating protein gelonin;PDBTitle: structure of gap31
81 d1ndha2
not modelled
6.9
22
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: Reductases
82 c6fhnA_
not modelled
6.9
20
PDB header: hydrolaseChain: A: PDB Molecule: protein;PDBTitle: structural dynamics and catalytic properties of a multi-modular2 xanthanase (pt derivative)
83 c3i5tB_
not modelled
6.9
18
PDB header: transferaseChain: B: PDB Molecule: aminotransferase;PDBTitle: crystal structure of aminotransferase prk07036 from rhodobacter2 sphaeroides kd131
84 c2k2uB_
not modelled
6.8
33
PDB header: transcriptionChain: B: PDB Molecule: alpha trans-inducing protein;PDBTitle: nmr structure of the complex between tfb1 subunit of tfiih2 and the activation domain of vp16
85 c3fy6A_
not modelled
6.8
67
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: integron cassette protein;PDBTitle: structure from the mobile metagenome of v. cholerae. integron cassette2 protein vch_cass3
86 d1x7da_
not modelled
6.8
13
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Ornithine cyclodeaminase-like
87 c1tllA_
not modelled
6.7
23
PDB header: oxidoreductaseChain: A: PDB Molecule: nitric-oxide synthase, brain;PDBTitle: crystal structure of rat neuronal nitric-oxide synthase2 reductase module at 2.3 a resolution.
88 c6m9sC_
not modelled
6.6
12
PDB header: oxidoreductaseChain: C: PDB Molecule: sznf;PDBTitle: crystal structure of semet sznf from streptomyces achromogenes var.2 streptozoticus nrrl 2697
89 d1qtma1
not modelled
6.5
44
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: DnaQ-like 3'-5' exonuclease
90 d2c7fa1
not modelled
6.5
21
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: Composite domain of glycosyl hydrolase families 5, 30, 39 and 51
91 c4nn3A_
not modelled
6.5
11
PDB header: transport proteinChain: A: PDB Molecule: trap dicarboxylate transporter, dctp subunit;PDBTitle: crystal structure of a trap periplasmic solute binding protein from2 desulfovibrio salexigens (desal_2161), target efi-510109, with bound3 orotic acid
92 c3b5oA_
not modelled
6.4
36
PDB header: oxidoreductaseChain: A: PDB Molecule: cadd-like protein of unknown function;PDBTitle: crystal structure of a cadd-like protein of unknown function2 (npun_f6505) from nostoc punctiforme pcc 73102 at 1.35 a resolution
93 c6cboB_
not modelled
6.4
16
PDB header: transferaseChain: B: PDB Molecule: c-6' aminotransferase;PDBTitle: x-ray structure of genb1 from micromonospora echinospora in complex2 with neamine and plp (as the external aldimine)
94 c1zrtD_
not modelled
6.3
39
PDB header: oxidoreductase/metal transportChain: D: PDB Molecule: cytochrome c1;PDBTitle: rhodobacter capsulatus cytochrome bc1 complex with2 stigmatellin bound
95 d1abra_
not modelled
6.3
19
Fold: Ribosome inactivating proteins (RIP)Superfamily: Ribosome inactivating proteins (RIP)Family: Plant cytotoxins
96 c2ke4A_
not modelled
6.3
75
PDB header: membrane proteinChain: A: PDB Molecule: cdc42-interacting protein 4;PDBTitle: the nmr structure of the tc10 and cdc42 interacting domain2 of cip4
97 c2bn5B_
not modelled
6.2
63
PDB header: nuclear proteinChain: B: PDB Molecule: u1 small nuclear ribonucleoprotein 70 kda;PDBTitle: p-element somatic inhibitor protein complex with u1-70k2 proline-rich peptide
98 d1b93a_
not modelled
6.1
29
Fold: Methylglyoxal synthase-likeSuperfamily: Methylglyoxal synthase-likeFamily: Methylglyoxal synthase, MgsA
99 c6ajrA_
not modelled
6.1
18
PDB header: dna binding proteinChain: A: PDB Molecule: uracil dna glycosylase superfamily protein;PDBTitle: complex form of uracil dna glycosylase x and uracil