| 1 |
|
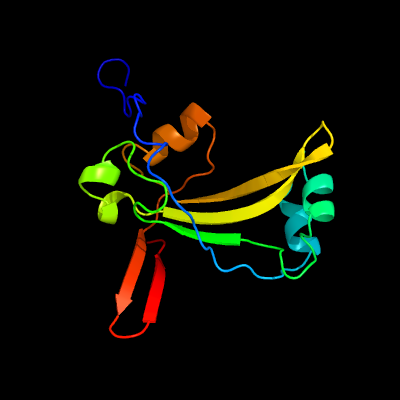
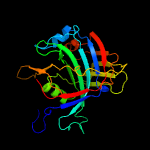
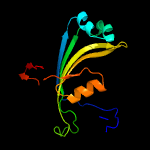
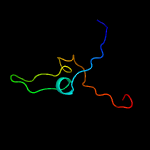
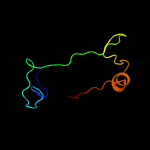
PDB 3bgt chain D
Region: 21 - 160
Aligned: 124
Modelled: 139
Confidence: 99.7%
Identity: 16%
PDB header:lyase
Chain: D: PDB Molecule:probable acetoacetate decarboxylase;
PDBTitle: structural studies of acetoacetate decarboxylase
Phyre2
| 2 |
|
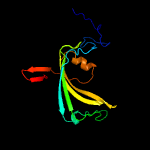
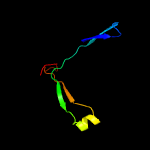
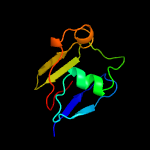
PDB 3bh2 chain C
Region: 13 - 160
Aligned: 133
Modelled: 139
Confidence: 99.6%
Identity: 12%
PDB header:lyase
Chain: C: PDB Molecule:acetoacetate decarboxylase;
PDBTitle: structural studies of acetoacetate decarboxylase
Phyre2
| 3 |
|
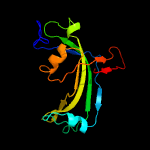
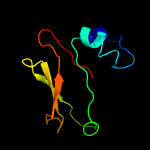
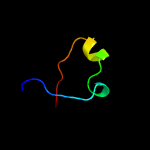
PDB 4jme chain A
Region: 18 - 252
Aligned: 226
Modelled: 235
Confidence: 99.4%
Identity: 17%
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein mppr;
PDBTitle: enduracididine biosynthesis enzyme mppr complexed with 2-keto-2 enduracididine
Phyre2
| 4 |
|
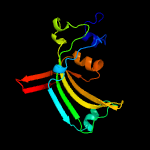
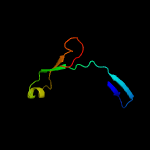
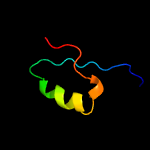
PDB 3c8w chain A domain 1
Region: 8 - 160
Aligned: 137
Modelled: 143
Confidence: 99.0%
Identity: 13%
Fold: Acetoacetate decarboxylase-like
Superfamily: Acetoacetate decarboxylase-like
Family: Acetoacetate decarboxylase-like
Phyre2
| 5 |
|
PDB 4zbt chain B
Region: 21 - 160
Aligned: 134
Modelled: 140
Confidence: 99.0%
Identity: 15%
PDB header:lyase
Chain: B: PDB Molecule:acetoacetate decarboxylase;
PDBTitle: streptomyces bingchenggensis aldolase-dehydratase in schiff base2 complex with pyruvate
Phyre2
| 6 |
|
PDB 5upb chain A
Region: 11 - 159
Aligned: 134
Modelled: 149
Confidence: 98.9%
Identity: 14%
PDB header:lyase
Chain: A: PDB Molecule:acetoacetate decarboxylase;
PDBTitle: swit_4259, an acetoacetate decarboxylase-like enzyme from sphingomonas2 wittichii rw1
Phyre2
| 7 |
|
PDB 3cmb chain A domain 1
Region: 21 - 159
Aligned: 127
Modelled: 138
Confidence: 98.6%
Identity: 13%
Fold: Acetoacetate decarboxylase-like
Superfamily: Acetoacetate decarboxylase-like
Family: Acetoacetate decarboxylase-like
Phyre2
| 8 |
|
PDB 5jxp chain A
Region: 62 - 116
Aligned: 53
Modelled: 55
Confidence: 19.6%
Identity: 25%
PDB header:hydrolase
Chain: A: PDB Molecule:asp/glu-specific dipeptidyl-peptidase;
PDBTitle: crystal structure of porphyromonas endodontalis dpp11 in alternate2 conformation
Phyre2
| 9 |
|
PDB 1s1d chain A
Region: 223 - 254
Aligned: 32
Modelled: 31
Confidence: 14.0%
Identity: 31%
Fold: 5-bladed beta-propeller
Superfamily: Apyrase
Family: Apyrase
Phyre2
| 10 |
|
PDB 2yue chain A
Region: 48 - 130
Aligned: 76
Modelled: 83
Confidence: 12.6%
Identity: 17%
PDB header:rna binding protein
Chain: A: PDB Molecule:protein neuralized;
PDBTitle: solution structure of the neuz (nhr) domain in neuralized2 from drosophila melanogaster
Phyre2
| 11 |
|
PDB 1ttz chain A
Region: 44 - 57
Aligned: 14
Modelled: 14
Confidence: 10.7%
Identity: 14%
Fold: Thioredoxin fold
Superfamily: Thioredoxin-like
Family: Thioltransferase
Phyre2
| 12 |
|
PDB 3wol chain B
Region: 62 - 116
Aligned: 53
Modelled: 55
Confidence: 10.3%
Identity: 25%
PDB header:hydrolase
Chain: B: PDB Molecule:dipeptidyl aminopeptidase bii;
PDBTitle: crystal structure of the dap bii dipeptide complex i
Phyre2
| 13 |
|
PDB 5w6l chain A
Region: 68 - 112
Aligned: 43
Modelled: 45
Confidence: 9.5%
Identity: 12%
PDB header:toxin
Chain: A: PDB Molecule:rtx repeat-containing cytotoxin;
PDBTitle: crystal structure of rrsp, a martx toxin effector domain from vibrio2 vulnificus cmcp6
Phyre2
| 14 |
|
PDB 3jb9 chain K
Region: 207 - 216
Aligned: 10
Modelled: 10
Confidence: 7.5%
Identity: 40%
PDB header:rna binding protein/rna
Chain: K: PDB Molecule:pre-mrna-splicing factor prp5;
PDBTitle: cryo-em structure of the yeast spliceosome at 3.6 angstrom resolution
Phyre2
| 15 |
|
PDB 2j58 chain G
Region: 101 - 178
Aligned: 78
Modelled: 78
Confidence: 7.3%
Identity: 13%
PDB header:membrane protein
Chain: G: PDB Molecule:outer membrane lipoprotein wza;
PDBTitle: the structure of wza
Phyre2
| 16 |
|
PDB 5ijl chain A
Region: 131 - 156
Aligned: 26
Modelled: 26
Confidence: 6.8%
Identity: 19%
PDB header:transferase
Chain: A: PDB Molecule:dna polymerase ii large subunit;
PDBTitle: d-family dna polymerase - dp2 subunit (catalytic subunit)
Phyre2
| 17 |
|
PDB 3fxt chain B
Region: 110 - 143
Aligned: 32
Modelled: 34
Confidence: 5.7%
Identity: 16%
PDB header:gene regulation
Chain: B: PDB Molecule:nucleoside diphosphate-linked moiety x motif 6;
PDBTitle: crystal structure of the n-terminal domain of human nudt6
Phyre2
| 18 |
|
PDB 2b5d chain X domain 1
Region: 97 - 108
Aligned: 12
Modelled: 12
Confidence: 5.7%
Identity: 50%
Fold: immunoglobulin/albumin-binding domain-like
Superfamily: Families 57/38 glycoside transferase middle domain
Family: AmyC C-terminal domain-like
Phyre2
| 19 |
|
PDB 6gzf chain B
Region: 140 - 198
Aligned: 51
Modelled: 59
Confidence: 5.6%
Identity: 16%
PDB header:transferase
Chain: B: PDB Molecule:glutathione s-transferase;
PDBTitle: xi class gst from natrialba magadii
Phyre2
| 20 |
|
PDB 3bsu chain F
Region: 103 - 116
Aligned: 14
Modelled: 14
Confidence: 5.4%
Identity: 21%
PDB header:hydrolase/rna/dna
Chain: F: PDB Molecule:ribonuclease h1;
PDBTitle: hybrid-binding domain of human rnase h1 in complex with 12-2 mer rna/dna
Phyre2
| 21 |
|